This builds on Optimizing
To-do list¶
Can we diagnose overfitting?
Is there any structure in the weight matrices or tuning curves that depends on tau?
I wonder if a possible conclusion we might reach is that the network needs more hidden neurons when tau is smaller but is more robust / better performance? Interesting biological conclusion.
Can we figure out the strategy they’re using?
In the weight matrices below, the first half of the inputs are left ear, and the second half are right ear. That means that if one ear is largely negative and the other is largely positive, the network is doing a sort of ILD type computation? Which would be weird, but maybe suggests something is off in the stimulus generation if there are meaningful ILDs?
Other ideas are that it might just be using a mixed strategy, which if we could identify partial strategies we could quantify the mix.
Need to remember that is basically representing the distribution of ITD-sensitive cells (encoding strategy, compare to distribution of best delays) while is representing the decoding strategy (compare to Jeffress versus pattern match versus hemispheric).
# Global setup
run_sample_data = True # whether or not to run for a single example to test and show the code
run_sweep = False # whether or not to run a large sweep of parameters
stimulus_frequency = 50 # Hz
default_num_hidden = 8
default_num_classes = 180//15Imports and setup¶
import os
import pickle
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
from tqdm.notebook import tqdm
import torch
import torch.nn as nn
dtype = torch.float
# Check whether a GPU is available
if torch.cuda.is_available():
device = torch.device("cuda")
else:
device = torch.device("cpu")
# Not using Brian so we just use these constants to make equations look nicer below
second = 1
ms = 1e-3
Hz = 1Data generation¶
class ITDDataset(torch.utils.data.Dataset):
def __init__(self,
num_samples,
dt=1*ms, # large time step to make simulations run faster for tutorial
anf_per_ear=100, # repeats of each ear with independent noise
envelope_power=2, # higher values make sharper envelopes, easier
rate_max=600*Hz, # maximum Poisson firing rate
f=stimulus_frequency,# stimulus frequency
duration=.1*second, # stimulus duration
num_classes=default_num_classes, # number of IPD classes for discretisation
batch_size=128,
shuffle=True,
numpy_seed=32844534, # seed for numpy random number generator
):
self.num_samples = num_samples
self.dt = dt
self.anf_per_ear = anf_per_ear
self.envelope_power = envelope_power
self.rate_max = rate_max
self.f = f
self.duration = duration
self.duration_steps = int(np.round(duration/dt))
self.input_size = 2*anf_per_ear
self.num_classes = num_classes
self.batch_size = batch_size
self.shuffle = shuffle
if numpy_seed:
np.random.seed(numpy_seed)
self.ipd, self.spikes = self.random_ipd_input_signal()
self.data_loader = torch.utils.data.DataLoader(self, batch_size=batch_size, shuffle=shuffle)
# Generate an input signal (spike array) from array of true IPDs
def input_signal(self, ipd, generate_spikes=True):
num_samples = len(ipd)
T = np.arange(self.duration_steps)*self.dt # array of times
phi0 = np.random.rand()
phi = 2*np.pi*(self.f*T+phi0) # array of phases corresponding to those times with random offset
# each point in the array will have a different phase based on which ear it is
# and its delay
theta = np.zeros((num_samples, self.duration_steps, 2*self.anf_per_ear))
# for each ear, we have anf_per_ear different phase delays from 0 to pi/2 so
# that the differences between the two ears can cover the full range from -pi/2 to pi/2
phase_delays = np.linspace(0, np.pi/2, self.anf_per_ear)
# now we set up these theta to implement that. Some numpy vectorisation logic here which looks a little weird,
# but implements the idea in the text above.
theta[:, :, :self.anf_per_ear] = phi[np.newaxis, :, np.newaxis]+phase_delays[np.newaxis, np.newaxis, :]
theta[:, :, self.anf_per_ear:] = phi[np.newaxis, :, np.newaxis]+phase_delays[np.newaxis, np.newaxis, :]+ipd[:, np.newaxis, np.newaxis]
# now generate Poisson spikes at the given firing rate as in the previous notebook
if self.envelope_power==2:
# exact integral solution for envelope power 2
theta2 = theta + 2*np.pi*self.f*self.dt
rate_integ = self.rate_max/4*((-np.sin(2*theta2) - 8*np.cos(theta2) + np.sin(2*theta) + 8*np.cos(theta) + 12*np.pi*self.f*self.dt)/(8*np.pi*self.f))
else:
rate_integ = self.rate_max*self.dt*(0.5*(1+np.sin(theta)))**self.envelope_power
if generate_spikes:
spikes = np.random.rand(num_samples, self.duration_steps, 2*self.anf_per_ear)<rate_integ
return spikes
else:
return rate_integ
# Generate some true IPDs from U(-pi/2, pi/2) and corresponding spike arrays
def random_ipd_input_signal(self):
ipd = np.random.rand(self.num_samples)*np.pi-np.pi/2 # uniformly random in (-pi/2, pi/2)
spikes = self.input_signal(ipd)
ipd = torch.tensor(ipd, device=device, dtype=dtype)
spikes = torch.tensor(spikes, device=device, dtype=dtype)
return ipd, spikes
def discretise(self, ipds):
return ((ipds+np.pi/2)*self.num_classes/np.pi).long() # assumes input is tensor
def continuise(self, ipd_indices): # convert indices back to IPD midpoints
return (ipd_indices+0.5)/self.num_classes*np.pi-np.pi/2
def __len__(self):
return self.num_samples
def __getitem__(self, idx):
return self.spikes[idx, :, :], self.discretise(self.ipd[idx])
def visualise(self, w=4, h=2):
N = w*h
# Plot a few just to show how it looks
spikes = self.spikes.cpu()
plt.figure(figsize=(10, 4), dpi=100)
for i in range(N):
plt.subplot(h, w, i+1)
plt.imshow(spikes[i, :, :].T, aspect='auto', interpolation='nearest', cmap=plt.cm.gray_r)
plt.title(f'True IPD = {int(self.ipd[i]*180/np.pi)} deg')
if i>=4:
plt.xlabel('Time (steps)')
if i%4==0:
plt.ylabel('Input neuron index')
plt.tight_layout()
if run_sample_data:
train_data = ITDDataset(num_samples=128*128, batch_size=128)
test_data = ITDDataset(num_samples=32*128, batch_size=128)
test_data.visualise()
if not os.path.exists('figures_for_paper/basicmodel-stimuli-examples.eps'):
plt.savefig('figures_for_paper/basicmodel-stimuli-examples.eps', format='eps')
plt.savefig('figures_for_paper/basicmodel-stimuli-examples.tiff', format='tiff')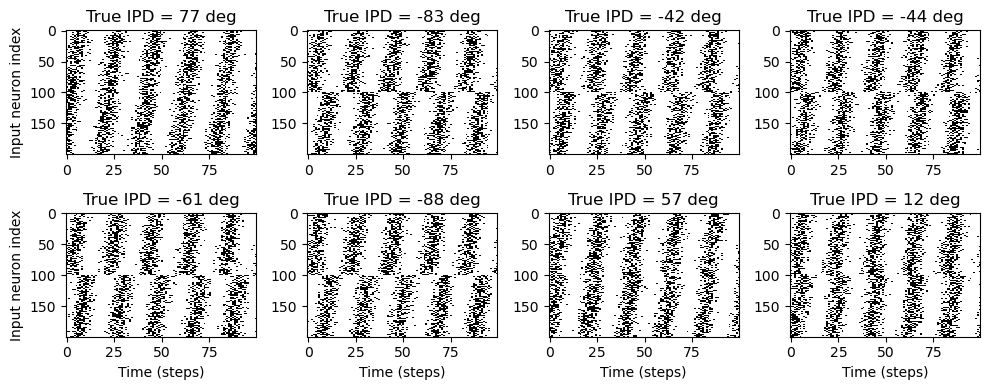
Model¶
Surrogate gradient function¶
class SurrGradSpike(torch.autograd.Function):
@staticmethod
def forward(ctx, input):
ctx.save_for_backward(input)
out = torch.zeros_like(input)
out[input > 0] = 1.0
return out
@staticmethod
def backward(ctx, grad_output):
input, = ctx.saved_tensors
# Original SPyTorch/SuperSpike gradient
# This seems to be a typo or error? But it works well
#grad = grad_output/(100*torch.abs(input)+1.0)**2
# Sigmoid
beta = 5
grad = grad_output*beta*torch.sigmoid(beta*input)*(1-torch.sigmoid(beta*input))
return grad
spike_fn = SurrGradSpike.applySpiking neural network¶
class ITDNet(nn.Module):
def __init__(self, dataset, num_hidden=default_num_hidden, tau=20*ms, dt=1*ms):
super(ITDNet, self).__init__()
self.input_size = dataset.input_size
self.num_classes = dataset.num_classes
self.num_hidden = num_hidden
self.tau = tau
self.dt = dt
self.init_weight_matrices()
# Weights and uniform weight initialisation
def init_weight_matrices(self):
# Input to hidden layer
self.W_ih = nn.Parameter(torch.empty((self.input_size, self.num_hidden), device=device, dtype=dtype, requires_grad=True))
fan_in, _ = nn.init._calculate_fan_in_and_fan_out(self.W_ih)
bound = 1 / np.sqrt(fan_in)
nn.init.uniform_(self.W_ih, -bound, bound)
# Hidden layer to output
self.W_ho = nn.Parameter(torch.empty((self.num_hidden, self.num_classes), device=device, dtype=dtype, requires_grad=True))
fan_in, _ = nn.init._calculate_fan_in_and_fan_out(self.W_ho)
bound = 1 / np.sqrt(fan_in)
nn.init.uniform_(self.W_ho, -bound, bound)
return self.W_ih, self.W_ho
def forward(self, input_spikes, apply_softmax=True):
batch_size, duration_steps, input_size = input_spikes.shape
if input_size!=self.input_size:
raise ValueError(f"Expected input size {self.input_size} but got {input_size}")
# First layer: input to hidden
v = torch.zeros((batch_size, self.num_hidden), device=device, dtype=dtype)
s = torch.zeros((batch_size, self.num_hidden), device=device, dtype=dtype)
s_rec = [s]
h = torch.einsum("abc,cd->abd", (input_spikes, self.W_ih))
alpha = np.exp(-self.dt/self.tau)
for t in range(duration_steps - 1):
new_v = (alpha*v + h[:, t, :])*(1-s) # multiply by 0 after a spike
s = spike_fn(v-1) # threshold of 1
v = new_v
s_rec.append(s)
s_rec = torch.stack(s_rec, dim=1)
# Second layer: hidden to output
v = torch.zeros((batch_size, self.num_classes), device=device, dtype=dtype)
s = torch.zeros((batch_size, self.num_classes), device=device, dtype=dtype)
v_rec = [v]
h = torch.einsum("abc,cd->abd", (s_rec, self.W_ho))
alpha = np.exp(-self.dt/self.tau)
for t in range(duration_steps - 1):
v = alpha*v + h[:, t, :]
v_rec.append(v)
v_rec = torch.stack(v_rec, dim=1)
# Return softmax of mean recorded membrane potential of output
m = torch.mean(v_rec, 1) # Mean across time dimension
if apply_softmax:
log_softmax_fn = nn.LogSoftmax(dim=1)
m = log_softmax_fn(m)
return m, s_rec
def train(self, nb_epochs=10, lr=0.01, fr_min=100*Hz, fr_max=200*Hz, pbar=None):
optimizer = torch.optim.Adam(self.parameters(), lr=lr)
loss_fn = nn.NLLLoss()
# training loop
loss_hist = []
initial_nll_loss_target = -np.log(1/train_data.num_classes)
if pbar is None:
print(f"Want loss for epoch 1 to be about {initial_nll_loss_target:.2f}, multiply m by constant to get this")
pbar = tqdm(total=nb_epochs)
close_pbar = True
else:
close_pbar = False
for e in range(nb_epochs):
local_loss = []
for spikes, ipds in train_data.data_loader:
# Run network
output, hidden_spikes = self(spikes)
fr = torch.sum(hidden_spikes, dim=(0, 1))/(train_data.batch_size*train_data.duration) # tensor of length num_hidden
reg_loss = torch.mean(initial_nll_loss_target*(torch.nn.functional.relu(fr-fr_min)/(fr_max-fr_min))**2)/self.num_hidden
loss = loss_fn(output, ipds)+reg_loss
local_loss.append(loss.item())
# Update gradients
loss.backward()
optimizer.step()
optimizer.zero_grad()
loss_hist.append(np.mean(local_loss))
pbar.update(1)
pbar.set_postfix(epoch=e, loss=loss_hist[-1])
if close_pbar:
pbar.close()
return loss_histTraining¶
if run_sample_data:
# net = ITDNet(train_data, tau=2*ms)
#loss_hist = net.train(nb_epochs=10, lr=0.01) # quick training (a few minutes)
#loss_hist = net.train(nb_epochs=100, lr=0.001) # long training for best results (about 30m)
#torch.save(net, 'saved_trained_models/itdnet-for-paper.pt')
net = torch.load('saved_trained_models/itdnet-for-paper.pt')
# Plot the loss function over time
# plt.plot(loss_hist)
# plt.xlabel('Epoch')
# plt.ylabel('Loss')
# plt.tight_layout()Analysis of trained network¶
class AnalysisResults:
def __init__(self, model, dataset, label, record_spikes=False):
self.model = model
self.dataset = dataset
self.label = label
self.record_spikes = record_spikes
# compute results on test dataset
all_output = []
all_ipd_idx_true = []
all_spikes = []
all_hidden_fr = []
for spikes, ipds in dataset.data_loader:
with torch.no_grad():
output, spikes_rec = model(spikes, apply_softmax=False)
# spikes_rec has shape (batch_size, duration_steps, num_hidden)
all_output.append(output)
all_ipd_idx_true.append(ipds)
all_hidden_fr.append(torch.sum(spikes_rec, dim=1)/self.dataset.duration)
if record_spikes:
for b in range(spikes_rec.shape[0]): # for each batch
time_idx, hidden_idx = torch.nonzero(spikes_rec[b, :, :]).detach().cpu().numpy().T
all_spikes.append((time_idx, hidden_idx))
self.all_output = all_output = torch.cat(all_output, 0).detach().cpu().numpy()
self.all_ipd_idx_est = all_ipd_idx_est = np.argmax(all_output, 1)
self.all_ipd_idx_true = all_ipd_idx_true = torch.cat(all_ipd_idx_true, 0).detach().cpu().numpy()
self.all_ipd_est = all_ipd_est = dataset.continuise(all_ipd_idx_est)
self.all_ipd_true = all_ipd_true = dataset.continuise(all_ipd_idx_true)
self.all_spikes= all_spikes
self.all_hidden_fr = torch.cat(all_hidden_fr, 0).detach().cpu().numpy()
# confusion matrix
self.confusion = confusion = np.zeros((dataset.num_classes, dataset.num_classes))
for i, j in zip(all_ipd_idx_true, all_ipd_idx_est):
confusion[j, i] += 1
confusion /= np.sum(confusion, axis=0)[np.newaxis, :]
# tuning curves of output neurons
def compute_tuning_curves(X): # X should have shape (num_samples, num_neurons)
num_neurons = X.shape[1]
tuning_curve_mean = np.zeros((dataset.num_classes, num_neurons))
tuning_curve_std = np.zeros((dataset.num_classes, num_neurons))
for i in range(dataset.num_classes):
o = X[all_ipd_idx_true==i, :]
tuning_curve_mean[i, :] = np.mean(o, axis=0)
tuning_curve_std[i, :] = np.std(o, axis=0)
return tuning_curve_mean, tuning_curve_std
self.tuning_curves = {
'output': compute_tuning_curves(all_output),
'hidden': compute_tuning_curves(self.all_hidden_fr),
}
# accuracy
self.accuracy = ((all_ipd_idx_est==all_ipd_idx_true)*1.0).mean()
self.mean_abs_error_deg = np.mean(abs(all_ipd_true-all_ipd_est)*180/np.pi)
def print_text(self):
print(f"Accuracy on {self.label} set = {self.accuracy*100:.1f}%, mean absolute error = {self.mean_abs_error_deg:.1f} deg")
return self
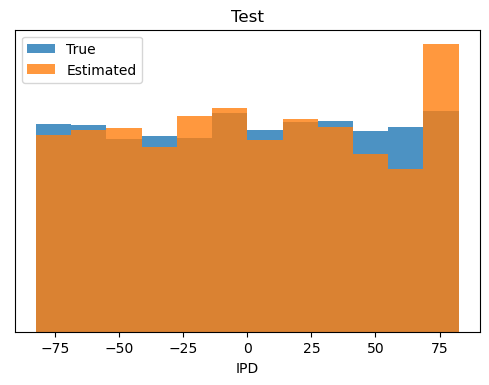
def plot_histograms(self):
plt.hist(self.all_ipd_true*180/np.pi, bins=self.dataset.num_classes, label='True', alpha=0.8)
plt.hist(self.all_ipd_est*180/np.pi, bins=self.dataset.num_classes, label='Estimated', alpha=0.8)
plt.xlabel("IPD")
plt.yticks([])
plt.legend(loc='best')
plt.title(self.label)
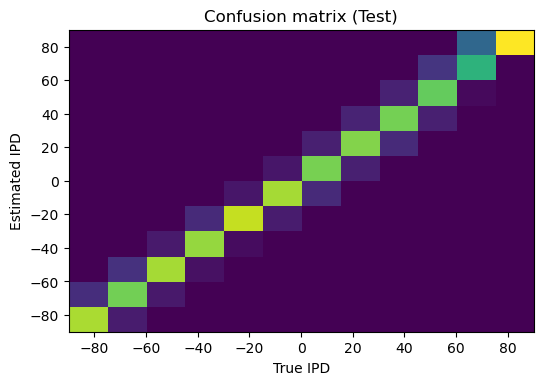
def plot_confusion(self):
plt.imshow(self.confusion, interpolation='nearest', aspect='auto', origin='lower', extent=(-90, 90, -90, 90))
plt.xlabel('True IPD')
plt.ylabel('Estimated IPD')
plt.title(f'Confusion matrix ({self.label})')
def plot_tuning_curves(self, name='output', fig=None, c='k', label=None, normalise=False, reorder=None):
tuning_curve_mean, tuning_curve_std = self.tuning_curves[name]
if normalise:
if normalise=='individual':
mins = tuning_curve_mean.min(axis=0)[None, :]
maxs = tuning_curve_mean.max(axis=0)[None, :]
elif normalise=='population':
mins = tuning_curve_mean.min()[None, None]
maxs = tuning_curve_mean.max()[None, None]
else:
raise ValueError("normalise should be 'individual' or 'population'")
tuning_curve_mean = (tuning_curve_mean-mins)/(maxs-mins)
tuning_curve_std = tuning_curve_std/(maxs-mins)
vertical_label = f'FR (norm={normalise})'
else:
mins = 0
maxs = 1
vertical_label = 'Firing rate (sp/s)'
if label is None:
label = self.label
if fig is None:
plt.figure(figsize=(12, 6), dpi=80)
else:
plt.figure(fig)
num_neurons = tuning_curve_mean.shape[1]
all_output = (self.all_output-mins)/(maxs-mins)
for i in range(num_neurons):
plt.subplot(3, 4, i+1)
if reorder is not None:
i = reorder[i]
if name=='output':
plt.plot(self.all_ipd_true*180/np.pi, all_output[:, i], ',', c='lightgray')
plt.errorbar(self.dataset.continuise(np.arange(self.dataset.num_classes))*180/np.pi, tuning_curve_mean[:, i], tuning_curve_std[:, i]*1.96, c=c, label=label)
if name=='output':
plt.axvline(self.dataset.continuise(i)*180/np.pi, ls='--', c='r', label='True IPD')
# if i//4==2:
plt.xlabel('IPD (deg)')
# if i%4==0:
if name=='output':
plt.ylabel(r'$\bar v$')
else:
plt.ylabel(vertical_label)
plt.ylim((tuning_curve_mean-1.96*tuning_curve_std).min(), (tuning_curve_mean+1.96*tuning_curve_std).max())
plt.suptitle(f"Tuning curves of {name} neurons")
plt.tight_layout()
def plot_weights(self, fig=None, reorder_by='ho'):
W_ih = self.model.W_ih.detach().cpu().numpy()
W_ho = self.model.W_ho.detach().cpu().numpy()
W_io = W_ih @ W_ho
if reorder_by=='ih':
# for each column of W_ih, compute the weighted mean and re-order according to that
A = np.arange(W_ih.shape[0]/2)
A = np.concatenate((A, -A))
A = A[:, None]
weighted_mean = np.mean((A*W_ih), axis=0)
# weighted_mean[np.max(np.abs(W_ih), axis=0)<.5] = np.inf
I = np.argsort(weighted_mean)
W_ih = W_ih[:, I]
W_ho = W_ho[I, :]
elif reorder_by=='ho':
# reorder by rows of W_ho
A = np.arange(W_ho.shape[1])[None, :]
weighted_mean = np.mean((A*W_ho), axis=1)
I = np.argsort(-weighted_mean)
W_ih = W_ih[:, I]
W_ho = W_ho[I, :]
def plot_matrix(W, x, y, t):
plt.imshow(W, interpolation='nearest', aspect='auto', origin='lower', vmin=-np.abs(W).max(), vmax=np.abs(W).max(), cmap='RdBu')
plt.ylabel(y)
plt.xlabel(x)
plt.title(t)
plt.colorbar(label="Weight")
if fig is None:
plt.figure(figsize=(10, 3), dpi=100)
else:
plt.figure(fig)
plt.subplot(131)
plot_matrix(W_ih, 'Hidden layer neuron index', 'Input neuron index', r'$W_{ih}$'+(' (reordered)' if reorder_by else ''))
plt.subplot(132)
plot_matrix(W_ho, 'Output neuron index', 'Hidden layer neuron index', r'$W_{ho}$'+(' (reordered)' if reorder_by else ''))
plt.subplot(133)
plot_matrix(W_io, 'Output neuron index', 'Input neuron index', r'$W_{io} = W_{ih} \cdot W_{ho} $')
plt.tight_layout()
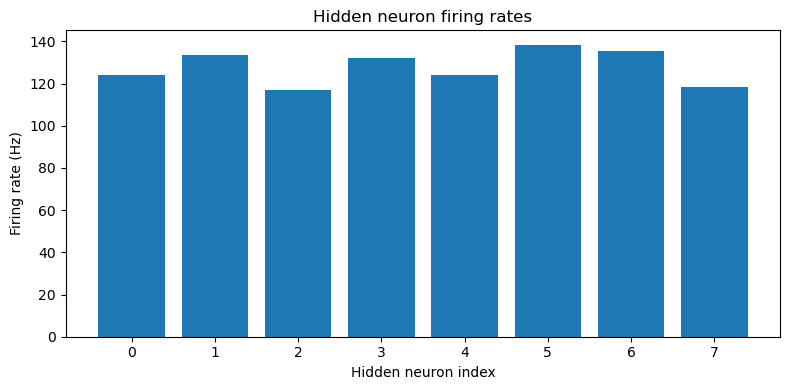
def plot_hidden_firing_rates(self):
plt.figure(figsize=(8, 4), dpi=100)
plt.bar(np.arange(self.model.num_hidden), self.all_hidden_fr.mean(axis=0))
plt.xlabel('Hidden neuron index')
plt.ylabel('Firing rate (Hz)')
plt.title('Hidden neuron firing rates')
plt.tight_layout()
def plot_rasters(self):
plt.figure(figsize=(10, 4), dpi=100)
for i in range(8):
plt.subplot(2, 4, i+1)
time_idx, hidden_idx = self.all_spikes[i]
plt.plot(time_idx*self.dataset.dt, hidden_idx, '.k', markersize=1)
plt.ylim(-1, self.model.num_hidden)
plt.xlim(0, self.dataset.duration)
plt.title(f'True IPD = {int(self.all_ipd_true[i]*180/np.pi)} deg')
if i>=4:
plt.xlabel('Time (steps)')
if i%4==0:
plt.ylabel('Hidden neuron index')
plt.tight_layout()
def visualise(self):
plt.figure(figsize=(6, 4), dpi=100)
# plt.subplot(121)
self.plot_histograms()
plt.figure(figsize=(6, 4), dpi=100)
# plt.subplot(122)
self.plot_confusion()
# plt.tight_layout()
if not os.path.exists('figures_for_paper/basicmodel-confusion.eps'):
plt.savefig('figures_for_paper/basicmodel-confusion.eps', format='eps', dpi=200)
plt.savefig('figures_for_paper/basicmodel-confusion.tiff', format='tiff', dpi=200)
self.plot_hidden_firing_rates()
if not os.path.exists('figures_for_paper/basicmodel-hidden-firing-rates.eps'):
plt.savefig('figures_for_paper/basicmodel-hidden-firing-rates.eps', format='eps', dpi=150)
plt.savefig('figures_for_paper/basicmodel-hidden-firing-rates.tiff', format='tiff', dpi=150)
if self.record_spikes:
self.plot_rasters()
self.plot_tuning_curves('output')
# if not os.path.exists('figures_for_paper/basicmodel-tuning-curves-output.eps'):
# plt.savefig('figures_for_paper/basicmodel-tuning-curves-output.eps', format='eps', dpi=150)
# plt.savefig('figures_for_paper/basicmodel-tuning-curves-output.tiff', format='tiff', dpi=150)
self.plot_tuning_curves('hidden')
# if not os.path.exists('figures_for_paper/basicmodel-tuning-curves-hidden.eps'):
# plt.savefig('figures_for_paper/basicmodel-tuning-curves-hidden.eps', format='eps', dpi=150)
# plt.savefig('figures_for_paper/basicmodel-tuning-curves-hidden.tiff', format='tiff', dpi=150)
self.plot_weights()
if not os.path.exists('figures_for_paper/basicmodel-weights.eps'):
plt.savefig('figures_for_paper/basicmodel-weights.eps', format='eps')
plt.savefig('figures_for_paper/basicmodel-weights.tiff', format='tiff')
return selfif run_sample_data:
print(f"Chance accuracy level: {100/train_data.num_classes:.1f}%")
# results_train = AnalysisResults(net, train_data, 'Train').print_text()
results_test = AnalysisResults(net, test_data, 'Test', record_spikes=True).print_text().visualise()Chance accuracy level: 8.3%
Accuracy on Test set = 82.6%, mean absolute error = 2.6 deg
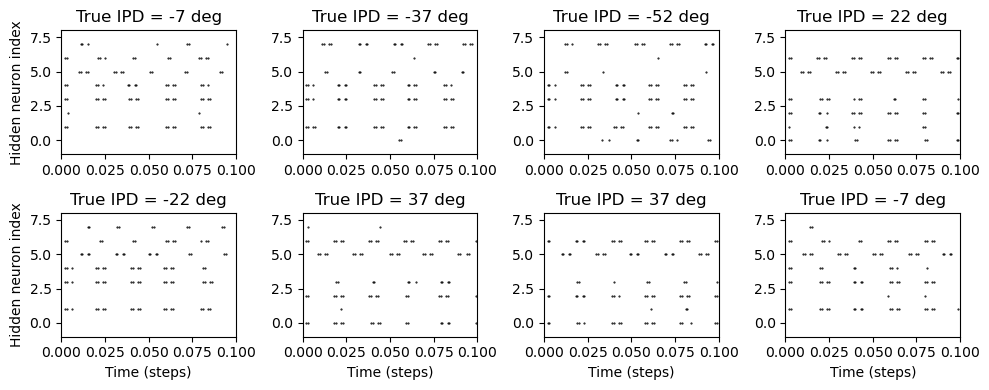
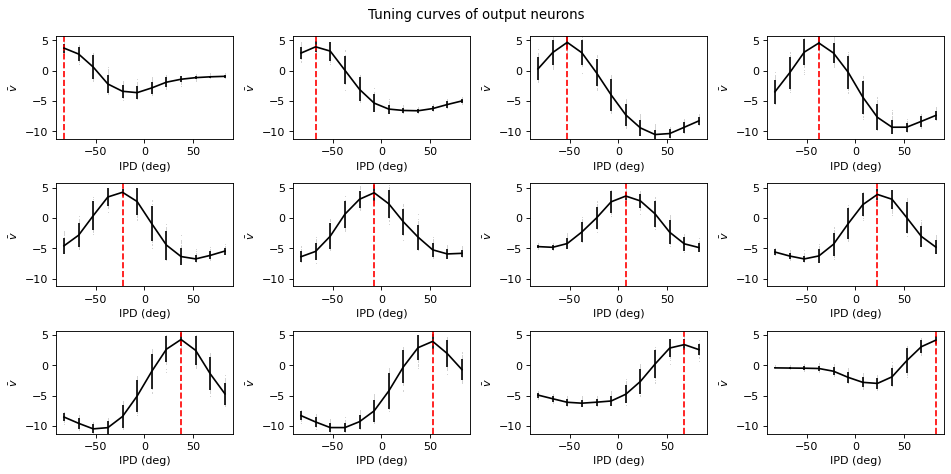
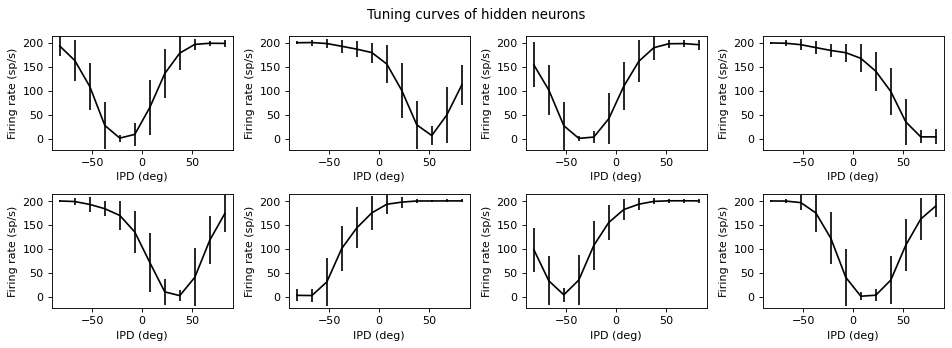
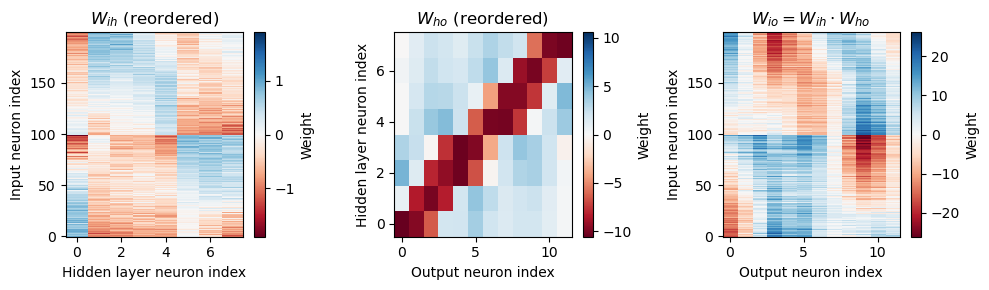
Understanding the solution found¶
Reorder matrices¶
Throughout this section we want the hidden units in order of their selectivity.
W_ih = net.W_ih.detach().cpu().numpy()
W_ho = net.W_ho.detach().cpu().numpy()
W_io = W_ih @ W_ho
# reorder by rows of W_ho
A = np.arange(W_ho.shape[1])[None, :]
weighted_mean = np.mean((A*W_ho), axis=1)
I_reorder = np.argsort(-weighted_mean)
W_ih = W_ih[:, I_reorder]
W_ho = W_ho[I_reorder, :]Fitting the hidden-to-output matrix to a simple function¶
The cell below shows the hidden->output matrix if the number of hidden neurons was the same as the number of output, and it was shifted. It shows that more or less the hidden->output matrix is just a sort of inverted Gaussian or maybe mexican hat around a moving centre.
W_ho_interp = torch.nn.functional.interpolate(torch.tensor(W_ho).reshape(1,1,8,12), size=(12, 12), mode='bicubic').detach().numpy()[0, 0, :, :]
I, J = np.meshgrid(np.arange(12), np.arange(12))
J = (I+J+6)%12
W_ho_interp = W_ho_interp[I, J]
plt.figure(figsize=(10, 3), dpi=100)
plt.subplot(1, 2, 1)
plt.imshow(W_ho_interp, interpolation='nearest', aspect='auto', origin='lower', vmin=-np.abs(W_ho).max(), vmax=np.abs(W_ho).max(), cmap='RdBu');
plt.subplot(1, 2, 2)
plt.plot(W_ho_interp.mean(axis=1))
plt.axhline(0, ls='--', c='k')
plt.tight_layout();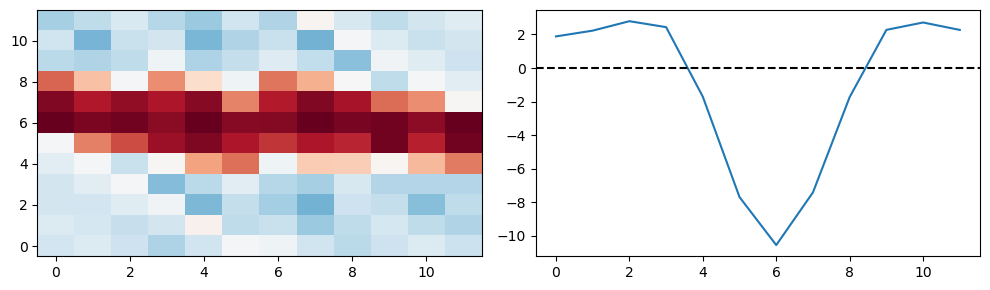
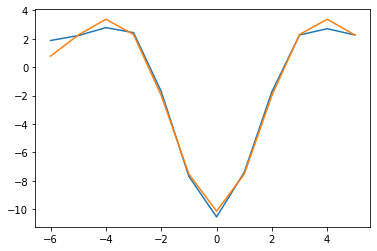
Let’s try to directly fit that with a Ricker (“mexican hat” function). It works well!
from scipy.optimize import curve_fit
psi = lambda t, a, b, sigma: a*(1-(t/sigma)**2)*np.exp(-t**2/(2*sigma**2))+b
t = np.arange(12)-6
y = W_ho_interp.mean(axis=1)
popt, pcov = curve_fit(psi, t, y, p0=[1, 0, 1])
print(popt)
plt.plot(t, y)
plt.plot(t, psi(t, *popt))[-9.36703502 -0.77569935 -2.22641301]
And now we fit the whole matrix with the same approach, looks good enough to try in the rate-based model.
psi = lambda t, a, b, sigma: a*(1-(t/sigma)**2)*np.exp(-t**2/(2*sigma**2))+b
H, O = np.meshgrid(np.arange(W_ho.shape[1]), np.arange(W_ho.shape[0]))
t = (O-W_ho.shape[0]*(H*1.0)/W_ho.shape[1]).flatten()
W_ho_flat = W_ho.flatten()
popt, pcov = curve_fit(psi, t, W_ho_flat, p0=[1, 0, 1])
W_ho_fit = psi(t, *popt).reshape(W_ho.shape)
plt.figure(figsize=(9, 4), dpi=100)
plt.subplot(1, 2, 1)
plt.imshow(W_ho, interpolation='nearest', aspect='auto', origin='lower', vmin=-np.abs(W_ho).max(), vmax=np.abs(W_ho).max(), cmap='RdBu')
plt.subplot(1, 2, 2)
plt.imshow(W_ho_fit.reshape(W_ho.shape), interpolation='nearest', aspect='auto', origin='lower', vmin=-np.abs(W_ho).max(), vmax=np.abs(W_ho).max(), cmap='RdBu')
plt.colorbar()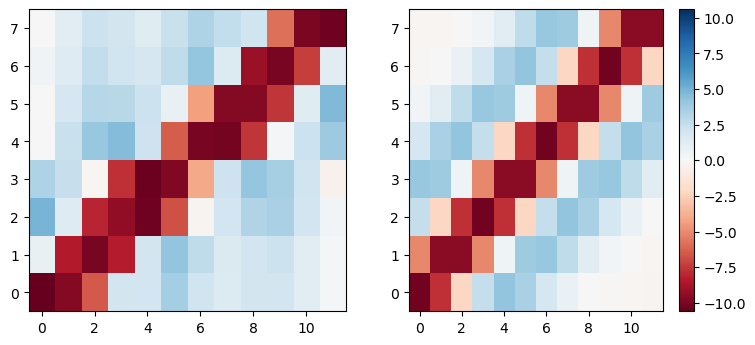
Fitting hidden neuron tuning curves to Gaussians¶
Fairly good fit.
tuning_curve_mean, _ = results_test.tuning_curves['hidden'] # (num_classes, num_neurons)
tuning_curve_mean = tuning_curve_mean[:, I_reorder]
ipds = np.linspace(-np.pi/2, np.pi/2, test_data.num_classes)
IPD, H = np.meshgrid(ipds, np.arange(net.num_hidden), indexing='ij')
t = -np.pi/2+H*np.pi/(net.num_hidden-1)-IPD
psi = lambda t, a, b, sigma: a*np.exp(-t**2/(2*sigma**2))+b
popt, pcov = curve_fit(psi, t.flatten(), tuning_curve_mean.flatten(), p0=[-200, 200, 1])
tuning_curve_fit = psi(t, *popt)
plt.figure(figsize=(10, 4), dpi=100)
for i in range(net.num_hidden):
plt.subplot(2, 4, i+1)
plt.plot(ipds*180/np.pi, tuning_curve_mean[:, i], label='Tuning curve')
plt.plot(ipds*180/np.pi, tuning_curve_fit[:, i], label='Fit')
plt.xlabel('IPD')
plt.ylabel('FR (sp/s)')
plt.legend(loc='best')
plt.tight_layout();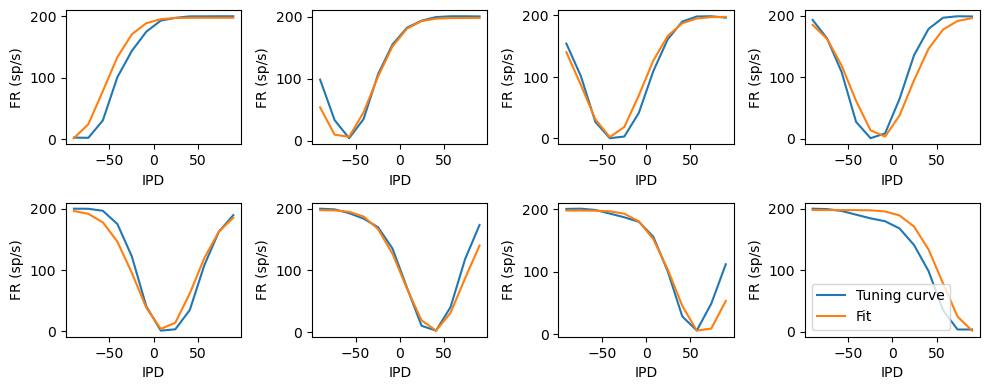
Approximating the firing rate curve with a simple function¶
In the cell below, we use the analytic function
for the firing rate of a leaky integrate-and-fire neuron under constant current with refractory period . We fit this to the firing rates we get for our neuron model experiencing a time-varying rate and get a pretty good fit. Note that because of the way we simulate it, we have an effective refractory period of and the currents have to be multiplied by .
# snn firing rates
rates_input = test_data.input_signal(ipds, generate_spikes=False)
x = np.einsum('ih,cti->cth', W_ih, rates_input)
tau = 2*ms
dt = 1*ms
alpha = np.exp(-dt/tau)
v = np.zeros((x.shape[0], x.shape[2]))
count = np.zeros_like(v)
for i in range(x.shape[1]):
v = alpha*v + x[:, i, :]
s = (v>1)*1.0
count += s
v[v>1] = 0
# now fit it
xclip = np.clip((tau/dt)*x, 1.001, np.inf)
y = (1/(dt+tau*np.log(xclip/(xclip-1))))*(x>1.001)
y = y.sum(axis=1)*dt
plt.plot(count.flatten(), y.flatten(), '.')
plt.plot(sorted(count.flatten()), sorted(count.flatten()), '--')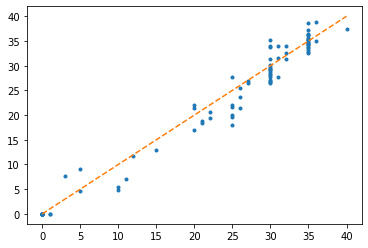
Predicting output curves based on rate-based model¶
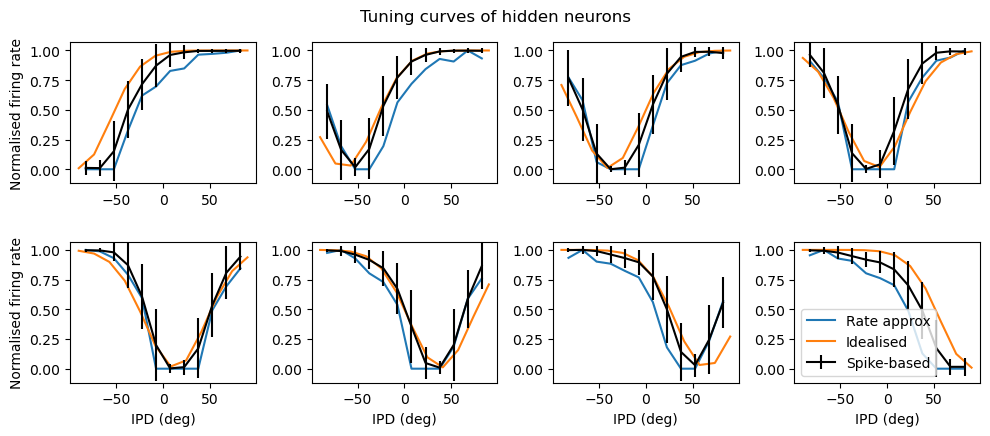
The idea of the cell below is to compute the mean output of each stage assuming no spiking. The fit is fairly good, suggesting that it’s not really using the spiking at all.
# Applying kernel makes no difference
# kernel_t = np.arange(-test_data.duration_steps+1, test_data.duration_steps)*test_data.dt
# kernel = (kernel_t>=0)*np.exp(-kernel_t/net.tau)
# plt.figure()
# plt.plot(kernel_t, kernel)
ipds = np.linspace(-np.pi/2, np.pi/2, test_data.num_classes)
rates_input = test_data.input_signal(ipds, generate_spikes=False)
# rates_hidden = np.clip(np.einsum('ih,cti->cth', W_ih, rates_input), 0, np.inf)
#sigmoid = lambda x: 1/(1+np.exp(-.4*x))
sigmoid_on_clipped = lambda x: 1/(dt+tau*np.log(x/(x-1)))
current_min = 1.001
sigmoid = lambda x: sigmoid_on_clipped(np.clip((tau/dt)*x, current_min, np.inf))*(x>current_min)
rates_hidden = sigmoid(np.einsum('ih,cti->cth', W_ih, rates_input))
# rates_hidden = np.einsum('ih,cti->cth', W_ih, rates_input) # this doesn't work well at all
# Applying kernel unnecessary for a good fit
# for c in range(rates_hidden.shape[0]):
# for h in range(rates_hidden.shape[2]):
# rates_hidden[c, :, h] = np.convolve(rates_hidden[c, :, h], kernel, mode='valid')
rates_output = np.einsum('ho,cth->cto', W_ho, rates_hidden)
rates_output_ricker = np.einsum('ho,cth->cto', W_ho_fit, rates_hidden)
rates_output_ricker_and_ideal = np.einsum('ho,ch->co', W_ho_fit, tuning_curve_fit)[:, None, :]
# Adding sigmoid here doesn't help which makes sense because these neurons are non-spiking
# sigmoid = lambda x: 1/(1+np.exp(-.1*x))
# rates_output = sigmoid(np.einsum('ho,cth->cto', W_ho, rates_hidden))
# rates_output_ricker = sigmoid(np.einsum('ho,cth->cto', W_ho_fit, rates_hidden))
# rates_input.shape # (num_classes, duration_steps, 2*anf_per_ear)
# plt.figure(figsize=(12, 6), dpi=100)
# for i in range(12):
# plt.subplot(3, 4, i+1)
# plt.imshow(rates_input[i, :, :].T, aspect='auto', interpolation='nearest', origin='lower', vmin=0)
# plt.suptitle('Mean input rates')
# plt.tight_layout()
# plt.figure(figsize=(12, 4), dpi=100)
# for i in range(8):
# plt.subplot(2, 4, i+1)
# plt.imshow(rates_hidden[i, :, :].T, aspect='auto', interpolation='nearest', origin='lower', vmin=0)
# plt.suptitle('Mean hidden rates')
# plt.tight_layout()
# plt.figure(figsize=(12, 6), dpi=100)
# for i in range(12):
# plt.subplot(3, 4, i+1)
# plt.imshow(rates_output[i, :, :].T, aspect='auto', interpolation='nearest', origin='lower', vmin=-np.abs(rates_output).max(), vmax=np.abs(rates_output).max(), cmap='RdBu')
# plt.suptitle('Mean output rates')
# plt.tight_layout()
# tuning curve comparison
fig = plt.figure(figsize=(10, 6), dpi=100)
for i in range(net.num_hidden):
plt.subplot(3, 4, i+1)
tc = rates_hidden[:, :, i].mean(axis=1)
tc = (tc-tc.min())/(tc.max()-tc.min())
plt.plot(results_test.dataset.continuise(np.arange(results_test.dataset.num_classes))*180/np.pi, tc, label='Rate approx')
plt.plot(ipds*180/np.pi, tuning_curve_fit[:, i]/np.max(tuning_curve_fit), label='Idealised')
results_test.plot_tuning_curves('hidden', normalise='population', label='Spike-based', reorder=I_reorder, fig=fig)
plt.legend(loc='best')
for i in range(4):
plt.subplot(3, 4, i+1)
plt.xlabel('')
for i in [1, 2, 3, 5, 6, 7]:
plt.subplot(3, 4, i+1)
plt.ylabel('')
for i in [0, 4]:
plt.subplot(3, 4, i+1)
plt.ylabel('Normalised firing rate')
plt.tight_layout()
if not os.path.exists('figures_for_paper/basicmodel-tuning-curves-hidden.eps'):
plt.savefig('figures_for_paper/basicmodel-tuning-curves-hidden.eps', format='eps')
plt.savefig('figures_for_paper/basicmodel-tuning-curves-hidden.tiff', format='tiff')
fig = plt.figure(figsize=(12, 6), dpi=100)
for i in range(12):
plt.subplot(3, 4, i+1)
tc = rates_output[:, :, i].mean(axis=1)
tc = (tc-tc.min())/(tc.max()-tc.min())
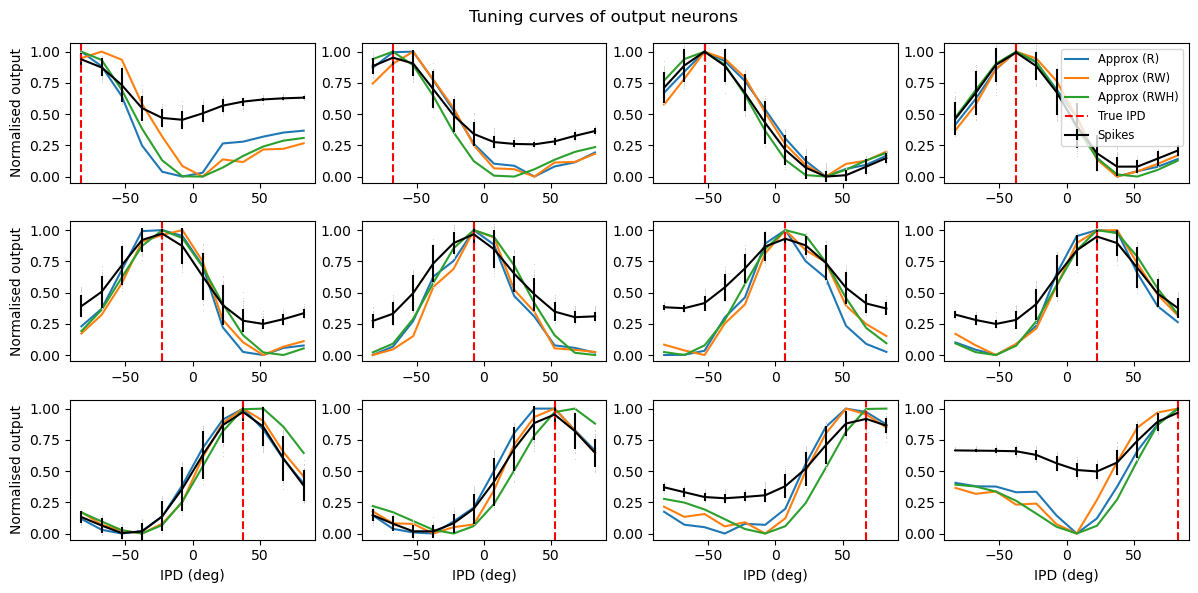
plt.plot(results_test.dataset.continuise(np.arange(results_test.dataset.num_classes))*180/np.pi, tc, label='Approx (R)')
tc = rates_output_ricker[:, :, i].mean(axis=1)
tc = (tc-tc.min())/(tc.max()-tc.min())
plt.plot(results_test.dataset.continuise(np.arange(results_test.dataset.num_classes))*180/np.pi, tc, label='Approx (RW)')
tc = rates_output_ricker_and_ideal[:, :, i].mean(axis=1)
tc = (tc-tc.min())/(tc.max()-tc.min())
plt.plot(results_test.dataset.continuise(np.arange(results_test.dataset.num_classes))*180/np.pi, tc, label='Approx (RWH)')
results_test.plot_tuning_curves('output', normalise='population', fig=fig, label='Spikes')
plt.subplot(3, 4, 4)
plt.legend(loc='best', fontsize='small')
for i in range(8):
plt.subplot(3, 4, i+1)
plt.xlabel('')
for i in [1, 2, 3, 5, 6, 7, 9, 10, 11]:
plt.subplot(3, 4, i+1)
plt.ylabel('')
for i in [0, 4, 8]:
plt.subplot(3, 4, i+1)
plt.ylabel('Normalised output')
plt.tight_layout();
if not os.path.exists('figures_for_paper/basicmodel-tuning-curves-output.eps'):
plt.savefig('figures_for_paper/basicmodel-tuning-curves-output.eps', format='eps')
plt.savefig('figures_for_paper/basicmodel-tuning-curves-output.tiff', format='tiff')The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
The PostScript backend does not support transparency; partially transparent artists will be rendered opaque.
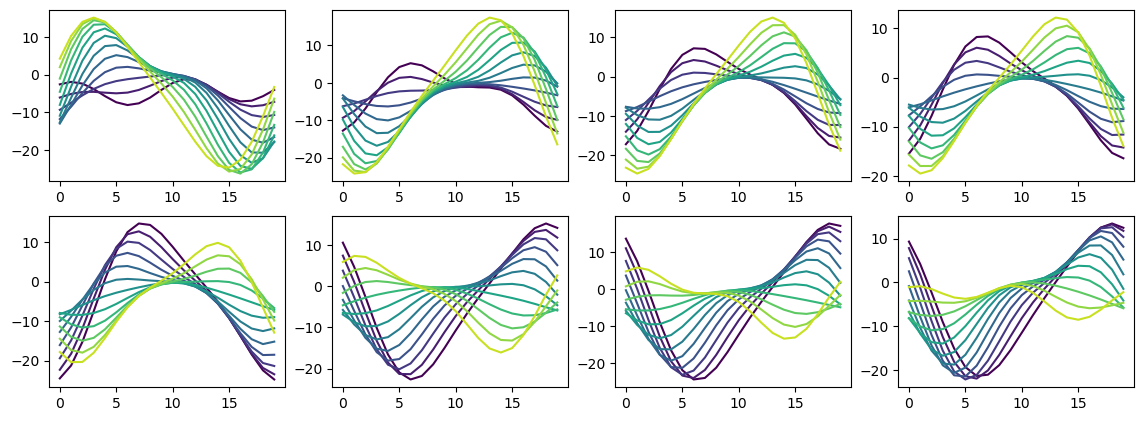
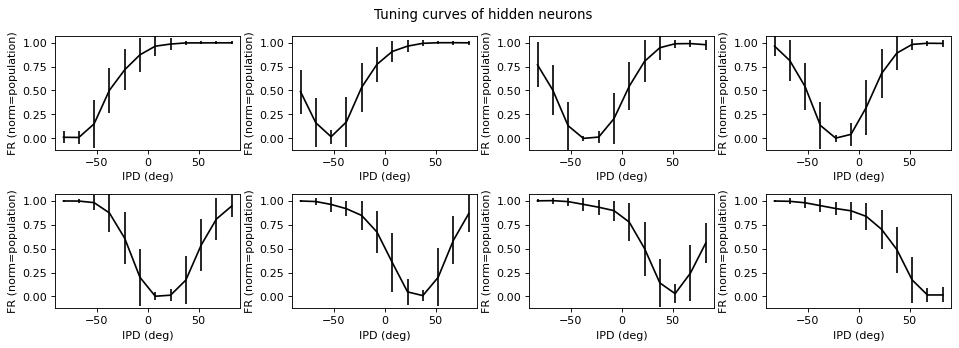
Understanding hidden unit tuning curve shape¶
Not clear there’s much to be got from this, it’s just a very overcomplete basis set.
single_phase_steps = int(1/(test_data.f*test_data.dt))
rates_hidden_L = np.einsum('ih,cti->cth', W_ih[:test_data.anf_per_ear, :], rates_input[:,:single_phase_steps,:test_data.anf_per_ear])
rates_hidden_R = np.einsum('ih,cti->cth', W_ih[test_data.anf_per_ear:, :], rates_input[:,:single_phase_steps,test_data.anf_per_ear:])
plt.figure(figsize=(14, 5), dpi=100)
for j in range(8):
plt.subplot(2, 4, j+1)
for i in range(test_data.num_classes):
plt.plot(rates_hidden_R[i, :, j], '-', c=plt.cm.viridis(i/test_data.num_classes))
# plt.plot(rates_hidden_L[0, :, j], '-', c=plt.cm.autumn(i/test_data.num_classes))
plt.plot(rates_hidden_L[0, :, j], '-k')
plt.figure(figsize=(14, 5), dpi=100)
for j in range(8):
plt.subplot(2, 4, j+1)
for i in range(test_data.num_classes):
# sigmoid = lambda x: 1/(1+np.exp(-0.3*x))
sigmoid = lambda x: x
plt.plot(sigmoid(rates_hidden_L[i, :, j]+rates_hidden_R[i, :, j]), '-', c=plt.cm.viridis(i/test_data.num_classes))
results_test.plot_tuning_curves('hidden', normalise='population', reorder=I_reorder)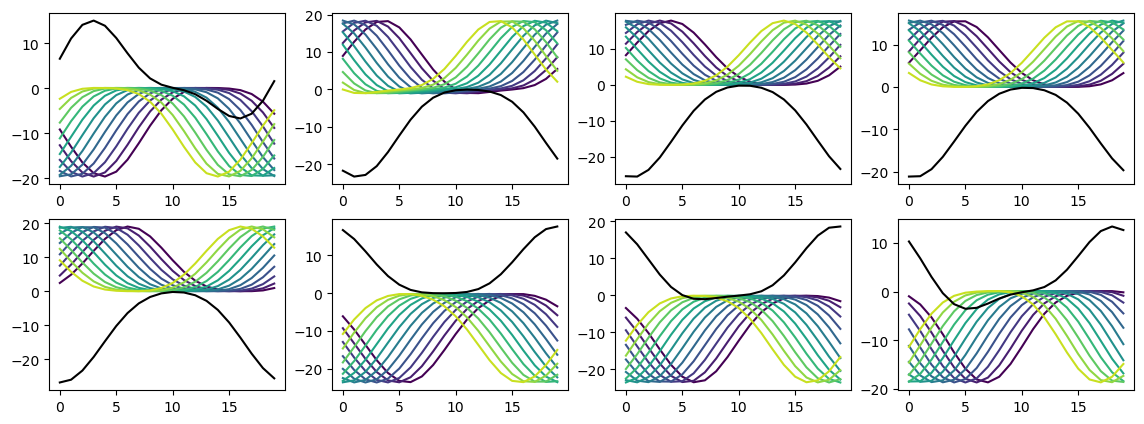
Varying ¶
Below we systematically vary the membrane time constant , saving results as we go, and then analyse the solutions found.
Run the simulations or load the saved data if available¶
if run_sweep:
# # Quick run settings for debugging
# tau_range = [2, 20]
# nb_epochs = 10
# num_hidden = 30
# Long run settings for detailed analysis
tau_range = [1, 2, 5, 10, 15, 20, 25, 30]
nb_epochs = 10 # actually seems to just start overfitting after 10 epochs
num_hidden = 30
# Common
num_classes = train_data.num_classes
# Run the simulations or load the data if saved
if not os.path.exists('tempdata'):
os.mkdir('tempdata')
pbar_steps = nb_epochs*len(tau_range)
nets = {}
results = {}
with tqdm(total=(nb_epochs+2)*len(tau_range)) as pbar:
for tau in tau_range:
fname = f'tempdata/itdnet_tau_{tau}_epochs_{nb_epochs}_hidden_{num_hidden}_classes_{num_classes}'
if os.path.exists(fname+'.pt'):
net = torch.load(fname+'.pt')
loss_hist = np.load(fname+'_loss.npy')
plt.plot(loss_hist, label=f'$\\{tau=}$ ms')
pbar.update(nb_epochs)
pbar.set_postfix(loaded=f'{tau=} ms')
else:
net = ITDNet(train_data, num_hidden=num_hidden, tau=tau*ms)
loss_hist = np.array(net.train(nb_epochs=nb_epochs, pbar=pbar))
np.save(fname+'_loss.npy', loss_hist)
torch.save(net, fname+'.pt')
# Plot the loss function over time
plt.plot(loss_hist, label=f'$\\{tau=}$ ms')
if os.path.exists(fname+'_results.pickle'):
with open(fname+'_results.pickle', 'rb') as f:
results_train, results_test = pickle.load(f)
pbar.update(2)
pbar.set_postfix(loaded_results=f'{tau=} ms')
else:
results_train = AnalysisResults(net, train_data, 'Train')
pbar.update(1)
pbar.set_postfix(results='train')
results_test = AnalysisResults(net, test_data, 'Test')
pbar.update(1)
pbar.set_postfix(results='test')
with open(fname+'_results.pickle', 'wb') as f:
pickle.dump((results_train, results_test), f)
results[tau, 'train'] = results_train
results[tau, 'test'] = results_test
print(f'{tau=} ms')
results_train.print_text()
results_test.print_text()
nets[tau] = net
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend(loc='best')
plt.tight_layout()Analyse the results¶
if run_sweep:
plt.figure(figsize=(8, 3), dpi=80)
plt.subplot(121)
for l in ['test', 'train']:
plt.plot(tau_range, [100*results[tau, l].accuracy for tau in tau_range], label=l)
plt.legend(loc='best')
plt.xlabel(r'$\tau$ (ms)')
plt.ylabel('Accuracy (%)')
plt.ylim(0, 100)
plt.subplot(122)
for l in ['test', 'train']:
plt.plot(tau_range, [results[tau, l].mean_abs_error_deg for tau in tau_range], label=l)
plt.xlabel(r'$\tau$ (ms)')
plt.ylabel('Mean absolute error (deg)')
plt.ylim(ymin=0)
plt.tight_layout()
fig = plt.figure(figsize=(12, 6), dpi=80)
for i, tau in enumerate(tau_range):
c = i/(len(tau_range)-1)
c = (c, c/2, 1-c)
results[tau, 'test'].plot_tuning_curves(fig=fig, c=c, label=f'$\\{tau=}$ ms')
plt.legend(loc='best')
for tau in tau_range:
results[tau, 'test'].plot_weights()
plt.suptitle(f'$\\{tau=}$ ms')
plt.tight_layout()