Code to generate parts of the figure in the main text.
Based on a pared-down version of the starting notebook.
import os
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpecSound localization stimuli¶
The following function creates a set of stimuli that can be used for training or testing.
We have two ears (0 and 1), and ear 1 will get a version of the signal delayed by an IPD we can write as in equations (ipd in code).
The basic signal is a sine wave made positive, so .
In addition, for each ear there will be neurons per ear (anf_per_ear because these are auditory nerve fibres).
Each neuron generates Poisson spikes at a certain firing rate, and these Poisson spike trains are independent.
In addition, since it is hard to train delays, we seed it with uniformly distributed delays from a minimum of 0 to a maximum of in each ear, so that the differences between the two ears can cover the range of possible IPDs ( to ). We do this directly by adding a phase delay to each neuron.
So for ear and neuron at time the angle .
Finally, we generate Poisson spike trains with a rate .
(
rate_max) is the maximum instantaneous firing rate, and(
envelope_power) is a constant that sharpens the envelope.
The higher and the easier the problem.
Here’s a picture of the architecture for the stimuli:

The functions below return two arrays ipd and spikes.
ipdis an array of lengthnum_samplesthat gives the true IPD, andspikesis an array of 0 (no spike) and 1 (spike) of shape(num_samples, duration_steps, 2*anf_per_ear), whereduration_stepsis the number of time steps there are in the stimulus.
# Not using Brian so we just use these constants to make equations look nicer below
second = 1
ms = 1e-3
Hz = 1
# Stimulus and simulation parameters
dt = 1*ms # large time step to make simulations run faster for tutorial
anf_per_ear = 100 # repeats of each ear with independent noise
envelope_power = 2 # higher values make sharper envelopes, easier
rate_max = 600*Hz # maximum Poisson firing rate
f = 20*Hz # stimulus frequency
duration = .1*second # stimulus duration
duration_steps = int(np.round(duration/dt))
input_size = 2*anf_per_ear
# Generate an input signal (spike array) from array of true IPDs
def input_signal(ipd):
num_samples = len(ipd)
T = np.arange(duration_steps)*dt # array of times
phi = 2*np.pi*(f*T+np.random.rand()) # array of phases corresponding to those times with random offset
# each point in the array will have a different phase based on which ear it is
# and its delay
theta = np.zeros((num_samples, duration_steps, 2*anf_per_ear))
# for each ear, we have anf_per_ear different phase delays from 0 to pi/2 so
# that the differences between the two ears can cover the full range from -pi/2 to pi/2
phase_delays = np.linspace(0, np.pi/2, anf_per_ear)
# now we set up these theta to implement that. Some numpy vectorisation logic here which looks a little weird,
# but implements the idea in the text above.
theta[:, :, :anf_per_ear] = phi[np.newaxis, :, np.newaxis] + phase_delays[np.newaxis, np.newaxis, :]
theta[:, :, anf_per_ear:] = phi[np.newaxis, :, np.newaxis] + phase_delays[np.newaxis, np.newaxis, :] + ipd[:, np.newaxis, np.newaxis]
# now generate Poisson spikes at the given firing rate
spikes = np.random.rand(num_samples, duration_steps, 2*anf_per_ear) < rate_max*dt*(0.5*(1+np.sin(theta)))**envelope_power
return spikes, phi
# Generate some true IPDs from U(-pi/2, pi/2) and corresponding spike arrays
def random_ipd_input_signal(num_samples):
ipd = np.random.rand(num_samples)*np.pi-np.pi/2 # uniformly random in (-pi/2, pi/2)
spikes = input_signal(ipd)[0]
return ipd, spikes
# Plot a few just to show how it looks
ipd, spikes = random_ipd_input_signal(8)
plt.figure(figsize=(10, 4), dpi=100)
for i in range(8):
plt.subplot(2, 4, i+1)
plt.imshow(spikes[i, :, :].T, aspect='auto', interpolation='nearest', cmap=plt.cm.gray_r)
plt.title(f'True IPD = {int(ipd[i]*180/np.pi)} deg')
if i>=4:
plt.xlabel('Time (steps)')
if i%4==0:
plt.ylabel('Input neuron index')
plt.tight_layout()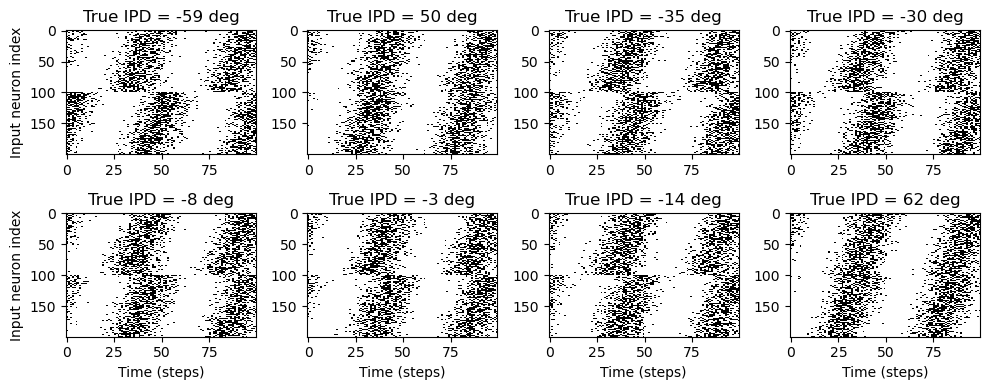
I think there is a little error here: the delays should have been subtracted instead of added (?).\
Take a peak of the first anf, 0 (say at IPD = 50°, t ≈ 45).
The other ANF’s of that ear (up to anf 99) should have that same peak later in time; but they have it earlier.
This is not really a problem (just relabel the neurons mentally, so that anf 99 receives the original signal, and anf 0 that signal delayed by 90°), it’s just a little confusing.
Select one IPD¶
ipd = np.pi * 0.2
ipd * 180/np.pi36.0np.random.seed(123414)
spikes, phi = input_signal(np.array([ipd]));
spikes.shape(1, 100, 200)(IPD, time, ANF)
def spiketrains(anf_nrs):
# Convert dense {0,1} array to spiketime array
spiketrain_list = []
for anf in anf_nrs:
spiketrain_list.append(np.nonzero(spikes[0, :, anf])[0])
return spiketrain_listdef rasterplot(trains, ax, title="Input spikes"):
ax.eventplot(trains, colors="black", lw=1, linelengths = 0.5);
ax.set(xlim=(0,100), xlabel="Time (steps)", ylabel="ANF number", title=title);
fig, ax = plt.subplots(figsize=(7,4))
rasterplot(spiketrains(np.arange(0,200,1)), ax)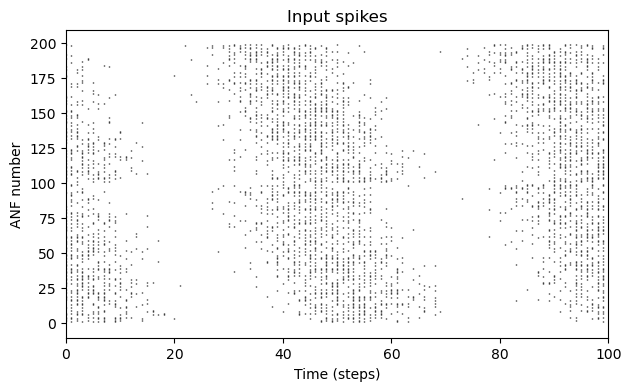
anfs = [0,99, 100,199]
fig, ax = plt.subplots(figsize=(7,2))
rasterplot(spiketrains(anfs), ax)
ax.set_yticks(np.arange(len(anfs)), anfs)
plt.savefig(Path("~/Desktop/spikes.svg").expanduser())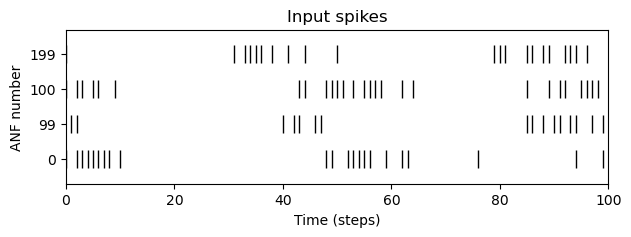
phase_delays = np.linspace(0, np.pi/2, anf_per_ear)
ipd_arr = ipd * np.ones_like(phase_delays)
theta = np.empty((len(phi), 2*anf_per_ear))
theta[:, :anf_per_ear] = phi[:,np.newaxis] + phase_delays[np.newaxis,:]
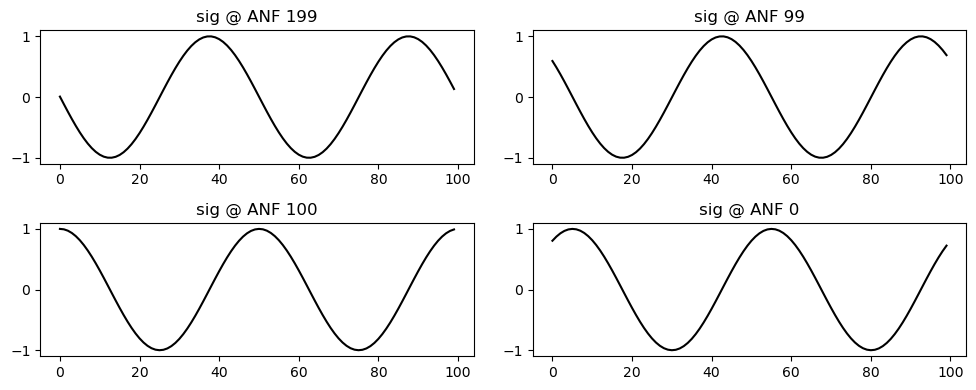
theta[:, anf_per_ear:] = phi[:,np.newaxis] + phase_delays[np.newaxis,:] + ipd_arr[np.newaxis,np.newaxis]fig, axs = plt.subplots(figsize=(10,4), nrows=2, ncols=2)
for (i,anf) in enumerate(anfs):
ax = axs.transpose().flatten()[3-i]
ax.plot(np.sin(theta[:,anf]), "k");
ax.set(title=f"sig @ ANF {anf}")
plt.tight_layout()
plt.savefig(Path("~/Desktop/sines.svg").expanduser())