Analysing Dale’s law and distribution of excitatory and inhibitory neurons¶
The first part is the standard quick start notebook, nothing has been changed.
Imports¶
%pip install tqdm
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors
import matplotlib.cm
import torch
import torch.nn as nn
from tqdm.auto import tqdm as pbar
dtype = torch.float
if torch.cuda.is_available():
device = torch.device("cuda")
else:
device = torch.device("cpu")Requirement already satisfied: tqdm in /Users/saraevers/miniconda3/envs/spikeloc/lib/python3.10/site-packages (4.64.1)
Note: you may need to restart the kernel to use updated packages.
Hyperparameters¶
# Constants
SECONDS = 1
MS = 1e-3
HZ = 1
DT = 1 * MS # large time step to make simulations run faster
ANF_PER_EAR = 100 # repeats of each ear with independent noise
DURATION = .1 * SECONDS # stimulus duration
DURATION_STEPS = int(np.round(DURATION / DT))
INPUT_SIZE = 2 * ANF_PER_EAR
# Training
LR = 0.01
N_EPOCHS = 50
batch_size = 64
n_training_batches = 64
n_testing_batches = 32
num_samples = batch_size*n_training_batches
# classes at 15 degree increments
NUM_CLASSES = 180 // 15
print(f'Number of classes = {NUM_CLASSES}')
# Network
NUM_HIDDEN = 30 # number of hidden units
TAU = 5 # membrane time constant
IE_RATIO = 0.5 # ratio of inhibitory:excitatory units (used if DALES_LAW = True). 0 = all excitatory, 1 = all inhibitory
DALES_LAW = True # When True, units will be only excitatory or inhibitory. When False, units will use both (like a normal ANN)
if DALES_LAW:
print('Using Dales Law')Number of classes = 12
Using Dales Law
Functions¶
Stimulus¶
def input_signal(ipd):
"""
Generate an input signal (spike array) from array of true IPDs
"""
envelope_power = 2 # higher values make sharper envelopes, easier
rate_max = 600 * HZ # maximum Poisson firing rate
stimulus_frequency = 20 * HZ
num_samples = len(ipd)
times = np.arange(DURATION_STEPS) * DT # array of times
phi = 2*np.pi*(stimulus_frequency * times + np.random.rand()) # array of phases corresponding to those times with random offset
# each point in the array will have a different phase based on which ear it is
# and its delay
theta = np.zeros((num_samples, DURATION_STEPS, 2*ANF_PER_EAR))
# for each ear, we have anf_per_ear different phase delays from to pi/2 so
# that the differences between the two ears can cover the full range from -pi/2 to pi/2
phase_delays = np.linspace(0, np.pi/2, ANF_PER_EAR)
# now we set up these theta to implement that. Some numpy vectorisation logic here which looks a little weird,
# but implements the idea in the text above.
theta[:, :, :ANF_PER_EAR] = phi[np.newaxis, :, np.newaxis]+phase_delays[np.newaxis, np.newaxis, :]
theta[:, :, ANF_PER_EAR:] = phi[np.newaxis, :, np.newaxis]+phase_delays[np.newaxis, np.newaxis, :]+ipd[:, np.newaxis, np.newaxis]
# now generate Poisson spikes at the given firing rate as in the previous notebook
spikes = np.random.rand(num_samples, DURATION_STEPS, 2*ANF_PER_EAR)<rate_max*DT*(0.5*(1+np.sin(theta)))**envelope_power
return spikes
def random_ipd_input_signal(num_samples, tensor=True):
"""
Generate the training data
Returns true IPDs from U(-pi/2, pi/2) and corresponding spike arrays
"""
ipd = np.random.rand(num_samples)*np.pi-np.pi/2 # uniformly random in (-pi/2, pi/2)
spikes = spikes_from_fixed_idp_input_signal(ipd, tensor)
if tensor:
ipd = torch.tensor(ipd, device=device, dtype=dtype)
return ipd, spikes
def spikes_from_fixed_idp_input_signal(ipd, tensor=True):
spikes = input_signal(ipd)
if tensor:
spikes = torch.tensor(spikes, device=device, dtype=dtype)
return spikes
def show_examples(shown=8):
ipd = np.linspace(-np.pi/2, np.pi/2, shown)
spikes = spikes_from_fixed_idp_input_signal(ipd, shown).cpu()
plt.figure(figsize=(10, 4), dpi=100)
for i in range(shown):
plt.subplot(2, shown // 2, i+1)
plt.imshow(spikes[i, :, :].T, aspect='auto', interpolation='nearest', cmap=plt.cm.gray_r)
plt.title(f'True IPD = {int(ipd[i]*180/np.pi)} deg')
if i>=4:
plt.xlabel('Time (steps)')
if i%4==0:
plt.ylabel('Input neuron index')
plt.tight_layout()
def data_generator(ipds, spikes):
perm = torch.randperm(spikes.shape[0])
spikes = spikes[perm, :, :]
ipds = ipds[perm]
n, _, _ = spikes.shape
n_batch = n//batch_size
for i in range(n_batch):
x_local = spikes[i*batch_size:(i+1)*batch_size, :, :]
y_local = ipds[i*batch_size:(i+1)*batch_size]
yield x_local, y_local
def discretise(ipds):
return ((ipds+np.pi/2) * NUM_CLASSES / np.pi).long() # assumes input is tensor
def continuise(ipd_indices): # convert indices back to IPD midpoints
return (ipd_indices+0.5) / NUM_CLASSES * np.pi - np.pi / 2SNN¶
def sigmoid(x, beta):
return 1 / (1 + torch.exp(-beta*x))
def sigmoid_deriv(x, beta):
s = sigmoid(x, beta)
return beta * s * (1 - s)
class SurrGradSpike(torch.autograd.Function):
@staticmethod
def forward(ctx, inp):
ctx.save_for_backward(inp)
out = torch.zeros_like(inp)
out[inp > 0] = 1.0
return out
@staticmethod
def backward(ctx, grad_output):
inp, = ctx.saved_tensors
sigmoid_derivative = sigmoid_deriv(inp, beta=5)
grad = grad_output*sigmoid_derivative
return grad
spike_fn = SurrGradSpike.apply
def membrane_only(input_spikes, weights, tau):
"""
:param input_spikes: has shape (batch_size, duration_steps, input_size)
:param weights: has shape (input_size, num_classes
:param tau:
:return:
"""
batch_size = input_spikes.shape[0]
assert len(input_spikes.shape) == 3
v = torch.zeros((batch_size, NUM_CLASSES), device=device, dtype=dtype)
v_rec = [v]
h = torch.einsum("abc,cd->abd", (input_spikes, weights))
alpha = np.exp(-DT / tau)
for t in range(DURATION_STEPS - 1):
v = alpha*v + h[:, t, :]
v_rec.append(v)
v_rec = torch.stack(v_rec, dim=1) # (batch_size, duration_steps, num_classes)
return v_rec
def layer1(input_spikes, w1, tau, sign1):
if DALES_LAW:
w1 = get_signed_weights(w1, sign1)
batch_size = input_spikes.shape[0]
# First layer: input to hidden
v = torch.zeros((batch_size, NUM_HIDDEN), device=device, dtype=dtype)
s = torch.zeros((batch_size, NUM_HIDDEN), device=device, dtype=dtype)
s_rec = [s]
h = torch.einsum("abc,cd->abd", (input_spikes, w1))
alpha = np.exp(-DT / tau)
for t in range(DURATION_STEPS - 1):
new_v = (alpha*v + h[:, t, :])*(1-s) # multiply by 0 after a spike
s = spike_fn(v-1) # threshold of 1
v = new_v
s_rec.append(s)
s_rec = torch.stack(s_rec, dim=1)
return s_rec
def layer2(s_rec, w2, tau, sign2):
"""Second layer: hidden to output"""
if DALES_LAW:
w2 = get_signed_weights(w2, sign2)
v_rec = membrane_only(s_rec, w2, tau=tau)
return v_rec
def snn(input_spikes, w1, w2, signs, tau=5*MS):
"""Run the simulation"""
s_rec = layer1(input_spikes, w1, tau, signs[0])
v_rec = layer2(s_rec, w2, tau, signs[1])
# Return recorded membrane potential of output
return v_recDale’s Law¶
def get_dales_mask(nb_inputs, nb_out, ie_ratio) :
d_mask = torch.ones(nb_inputs, nb_out)
#inhib_units = np.random.choice(nb_inputs, int(nb_inputs*ie_ratio), replace=False)
inhib_units = torch.arange(ie_ratio*nb_inputs, dtype=int)
d_mask[inhib_units, :] = -1
return d_mask
def init_weight_matrices(ie_ratio = 0.1):
"""Weights and uniform weight initialisation"""
# Input to hidden layer
w1 = nn.Parameter(torch.empty((INPUT_SIZE, NUM_HIDDEN), device=device, dtype=dtype, requires_grad=True))
fan_in, _ = nn.init._calculate_fan_in_and_fan_out(w1)
bound = 1 / np.sqrt(fan_in)
nn.init.uniform_(w1, -bound, bound)
# Hidden layer to output
w2 = nn.Parameter(torch.empty((NUM_HIDDEN, NUM_CLASSES), device=device, dtype=dtype, requires_grad=True))
fan_in, _ = nn.init._calculate_fan_in_and_fan_out(w2)
bound = 1 / np.sqrt(fan_in)
nn.init.uniform_(w2, -bound, bound)
#Get fixed signs for the weight, 90% excitatory
signs = [get_dales_mask(*w.shape, ie_ratio).to(w.device) for w in (w1, w2)]
return w1, w2, signs
def get_signed_weights(w, sign):
"""Get the signed value of the weight"""
# Note abs is in principle not differentiable.
# In practice, pytorch will set the derivative to 0 when the values are 0.
# (see https://discuss.pytorch.org/t/how-does-autograd-deal-with-non-differentiable-opponents-such-as-abs-and-max/34538)
# This has the adverse effect that, during training, if a synapse reaches 0,
# it is "culled" and can not be recovered.
# It should be possible to cheat here and either "wiggle" 0-valued synapses,
# or to override abs gradient to return a very small random number.
#TODO try ReLu or other activation
#TODO reproduce paper https://www.biorxiv.org/content/10.1101/2020.11.02.364968v2.full
# return torch.max(w, 0)*sign
return torch.abs(w)*signTraining¶
def train(w1, w2, signs, ipds, spikes, ipds_validation, spikes_validation, lr=0.01, n_epochs=30, tau = 5*MS):
"""
:param lr: learning rate
:return:
"""
# Optimiser and loss function
optimizer = torch.optim.Adam([w1, w2], lr=lr)
log_softmax_fn = nn.LogSoftmax(dim=1)
loss_fn = nn.NLLLoss()
loss_hist = []
val_loss_hist = []
best_loss = 1e10
val_loss_best_loss = 1e10
for e in pbar(range(n_epochs)):
local_loss = []
for x_local, y_local in data_generator(discretise(ipds), spikes):
# Run network
output = snn(x_local, w1, w2, signs, tau=tau)
# Compute cross entropy loss
m = torch.sum(output, 1)*0.01 # Sum time dimension
reg = 0
loss = loss_fn(log_softmax_fn(m), y_local) + reg
local_loss.append(loss.item())
# Update gradients
optimizer.zero_grad()
loss.backward()
optimizer.step()
loss_hist.append(np.mean(local_loss))
val_local_loss = []
for x_local, y_local in data_generator(discretise(ipds_validation), spikes_validation):
# Run network
output = snn(x_local, w1, w2, signs, tau=tau)
# Compute cross entropy loss
m = torch.sum(output, 1)*0.01 # Sum time dimension
val_loss = loss_fn(log_softmax_fn(m), y_local)
val_local_loss.append(val_loss.item())
val_loss_hist.append(np.mean(val_local_loss))
if np.mean(val_local_loss) < val_loss_best_loss:
val_loss_best_loss = np.mean(val_local_loss)
if DALES_LAW:
best_weights = get_signed_weights(w1, signs[0]), get_signed_weights(w2, signs[1]), signs
else:
best_weights = w1, w2, signs
#Early Stopping :
if torch.tensor(val_loss_hist[-100:]).argmin() == 0 and e>10:
print('Early Stop !')
return best_weights
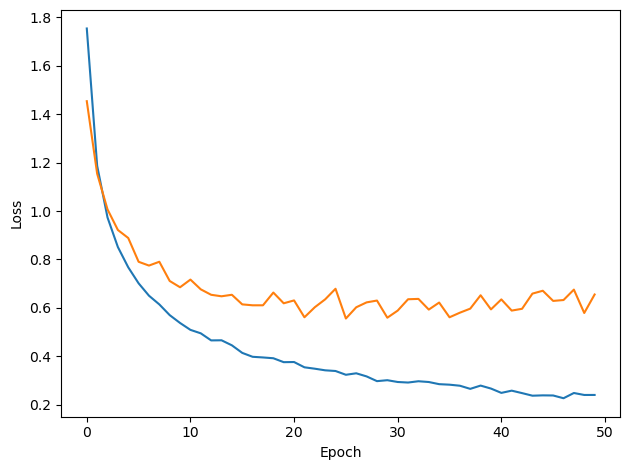
# Plot the loss function over time
plt.plot(loss_hist)
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.tight_layout()
plt.plot(val_loss_hist)
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.tight_layout()
if DALES_LAW:
return get_signed_weights(w1, signs[0]), get_signed_weights(w2, signs[1]), signs
else:
return w1, w2, signsTesting¶
def test_accuracy(ipds, spikes, run):
accs = []
ipd_true = []
ipd_est = []
confusion = np.zeros((NUM_CLASSES, NUM_CLASSES))
for x_local, y_local in data_generator(ipds, spikes):
y_local_orig = y_local
y_local = discretise(y_local)
output = run(x_local)
m = torch.sum(output, 1) # Sum time dimension
_, am = torch.max(m, 1) # argmax over output units
tmp = np.mean((y_local == am).detach().cpu().numpy()) # compare to labels
for i, j in zip(y_local.detach().cpu().numpy(), am.detach().cpu().numpy()):
confusion[j, i] += 1
ipd_true.append(y_local_orig.cpu().data.numpy())
ipd_est.append(continuise(am.detach().cpu().numpy()))
accs.append(tmp)
ipd_true = np.hstack(ipd_true)
ipd_est = np.hstack(ipd_est)
return ipd_true, ipd_est, confusion, accs
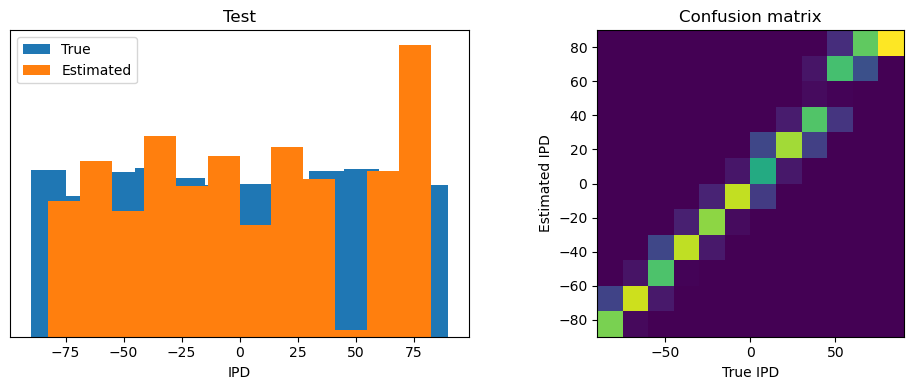
def report_accuracy(ipd_true, ipd_est, confusion, accs, label):
abs_errors_deg = abs(ipd_true-ipd_est)*180/np.pi
print()
print(f"{label} classifier accuracy: {100*np.mean(accs):.1f}%")
print(f"{label} absolute error: {np.mean(abs_errors_deg):.1f} deg")
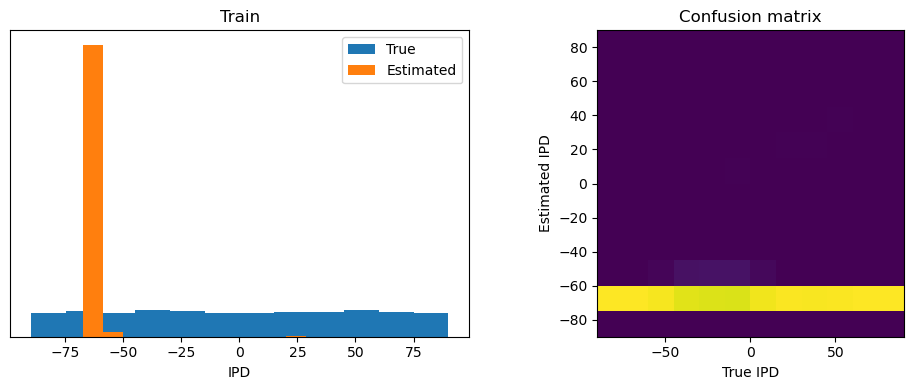
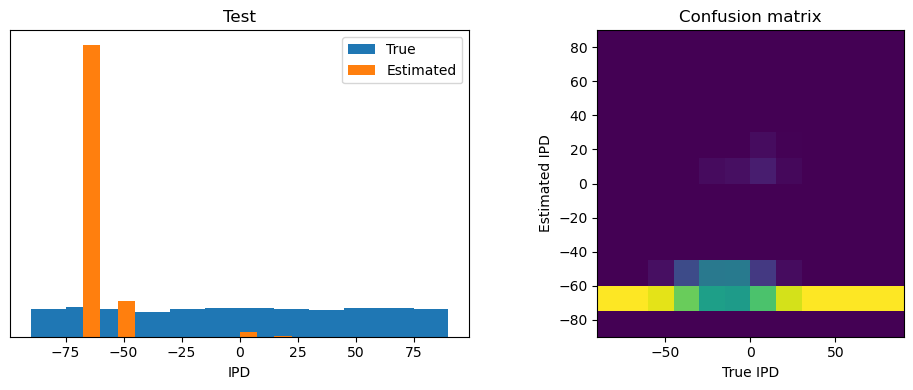
plt.figure(figsize=(10, 4), dpi=100)
plt.subplot(121)
plt.hist(ipd_true * 180 / np.pi, bins=NUM_CLASSES, label='True')
plt.hist(ipd_est * 180 / np.pi, bins=NUM_CLASSES, label='Estimated')
plt.xlabel("IPD")
plt.yticks([])
plt.legend(loc='best')
plt.title(label)
plt.subplot(122)
confusion /= np.sum(confusion, axis=0)[np.newaxis, :]
plt.imshow(confusion, interpolation='nearest', aspect='equal', origin='lower', extent=(-90, 90, -90, 90))
plt.xlabel('True IPD')
plt.ylabel('Estimated IPD')
plt.title('Confusion matrix')
plt.tight_layout()
def analyse_accuracy(ipds, spikes, run, label):
ipd_true, ipd_est, confusion, accs = test_accuracy(ipds, spikes, run)
report_accuracy(ipd_true, ipd_est, confusion, accs, label)
return 100*np.mean(accs)Train Network¶
# Generate the training data
w1, w2, signs = init_weight_matrices(ie_ratio=IE_RATIO)
ipds_training, spikes_training = random_ipd_input_signal(num_samples)
ipds_validation, spikes_validation = random_ipd_input_signal(num_samples)
# Train network
w1_trained, w2_trained, signs = train(w1, w2, signs, ipds_training, spikes_training, ipds_validation, spikes_validation, lr=LR, n_epochs=N_EPOCHS, tau=TAU*MS)Loading...
# Analyse
print(f"Chance accuracy level: {100 * 1 / NUM_CLASSES:.1f}%")
run_func = lambda x: snn(x, w1_trained, w2_trained, signs)
analyse_accuracy(ipds_training, spikes_training, run_func, 'Train')
ipds_test, spikes_test = random_ipd_input_signal(batch_size*n_testing_batches)
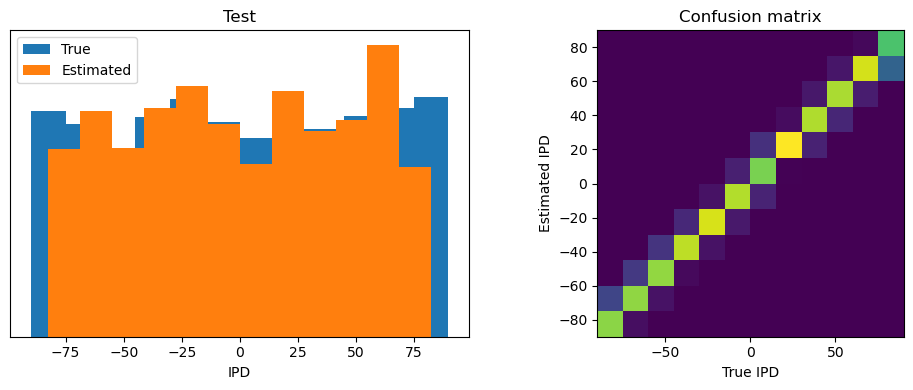
analyse_accuracy(ipds_test, spikes_test, run_func, 'Test')
Chance accuracy level: 8.3%
Train classifier accuracy: 92.7%
Train absolute error: 4.0 deg
Test classifier accuracy: 83.7%
Test absolute error: 4.4 deg
83.69140625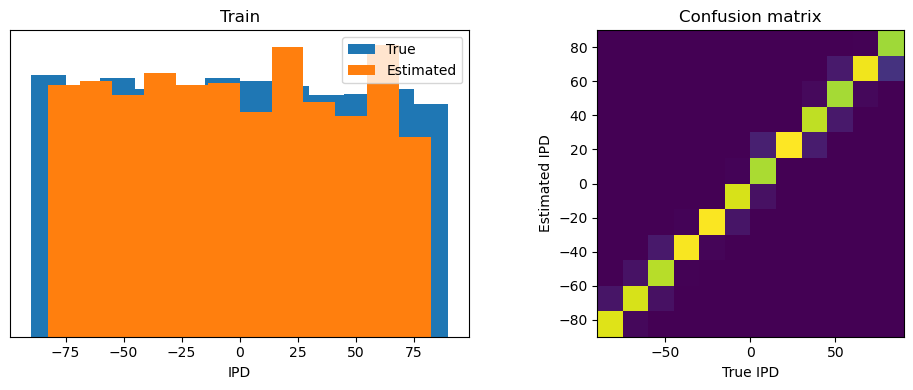
Analysis¶
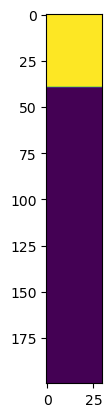
First, we visualise the distribution of excitatory and inhibitory neurons BEFORE and AFTER the learning
# Now we can remove all inhibitory connections to see what happens (yellow = 1, purple = 0)
f, axarr = plt.subplots(1,2)
axarr[0].imshow(w1.detach()>0)
axarr[0].set_title('Before learning')
axarr[0].set_ylabel('Neuron index')
axarr[0].set_xlabel('Hidden layer index')
axarr[1].imshow(w1_trained.detach()>0)
axarr[1].set_title('After learning')
axarr[1].set_ylabel('Neuron index')
axarr[1].set_xlabel('Hidden layer index')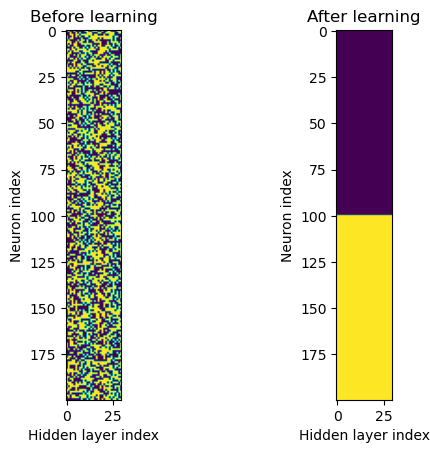
Interestingly, the same thing happens when we change the ratio of excitatory to inhibitory neurons, for example here to 80:20
# Change IE_RATIO
IE_RATIO = 0.2
# Generate the training data
w1, w2, signs = init_weight_matrices(ie_ratio=IE_RATIO)
ipds_training, spikes_training = random_ipd_input_signal(num_samples)
ipds_validation, spikes_validation = random_ipd_input_signal(num_samples)
# Train network
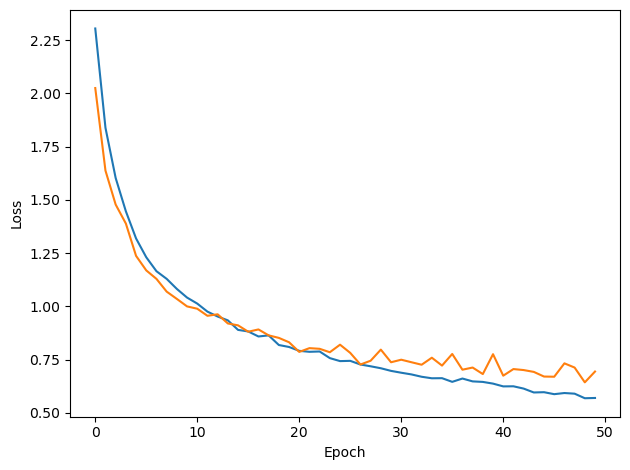
w1_trained, w2_trained, signs = train(w1, w2, signs, ipds_training, spikes_training, ipds_validation, spikes_validation, lr=LR, n_epochs=N_EPOCHS, tau=TAU*MS)Loading...
# Analyse
print(f"Chance accuracy level: {100 * 1 / NUM_CLASSES:.1f}%")
run_func = lambda x: snn(x, w1_trained, w2_trained, signs)
analyse_accuracy(ipds_training, spikes_training, run_func, 'Train')
ipds_test, spikes_test = random_ipd_input_signal(batch_size*n_testing_batches)
analyse_accuracy(ipds_test, spikes_test, run_func, 'Test')Chance accuracy level: 8.3%
Train classifier accuracy: 78.1%
Train absolute error: 5.3 deg
Test classifier accuracy: 70.6%
Test absolute error: 6.2 deg
70.60546875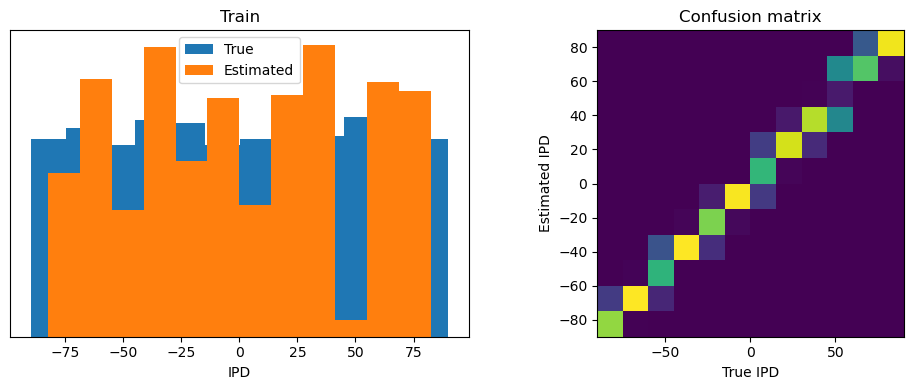
# Now we can remove all inhibitory connections to see what happens (yellow = 1, purple = 0)
f, axarr = plt.subplots(1,2)
axarr[0].imshow(w1.detach()>0)
axarr[0].set_title('Before learning')
axarr[0].set_ylabel('Neuron index')
axarr[0].set_xlabel('Hidden layer index')
axarr[1].imshow(w1_trained.detach()>0)
axarr[1].set_title('After learning')
axarr[1].set_ylabel('Neuron index')
axarr[1].set_xlabel('Hidden layer index')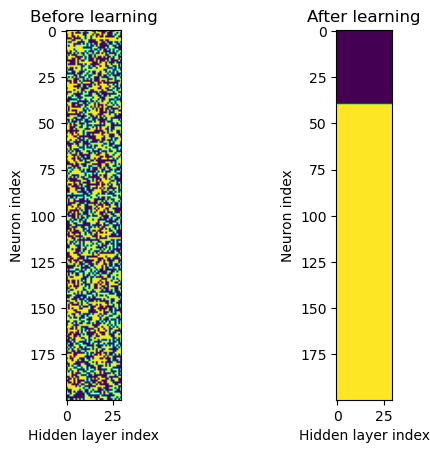
Again, we find that the learning results in a ‘phase seperation’ of inhibitory and excitatory neurons, which might be a mistake in the code or an interesting result.
Testing relevance of inhibitory neurons¶
Now I will set the inhibitory neurons equal to zero after the training, to see how relevant the inhibitory connections are for the performance of the network.
import random
w1_trained_new = w1_trained
for i in range(0,int(IE_RATIO*200)):
w1_trained_new[i,:] = w1_trained_new[i,:]*0
plt.imshow(w1_trained_new.detach() == 0)
run_func = lambda x: snn(x, w1_trained_new, w2_trained, signs)
analyse_accuracy(ipds_training, spikes_training, run_func, 'Train')
ipds_test, spikes_test = random_ipd_input_signal(batch_size*n_testing_batches)
analyse_accuracy(ipds_test, spikes_test, run_func, 'Test')
Train classifier accuracy: 8.6%
Train absolute error: 70.0 deg
Test classifier accuracy: 10.0%
Test absolute error: 68.0 deg
9.9609375