This notebook run the simulations reported in the paper, saves the results in a folder, and then plot the results to re-create the figures from the section “Contralateral glycinergic inhibition as a key factor in creating ITD sensitivity” of the paper.
Requirements¶
We tested the notebook with
Python version 3.12.3
NEST version 3.7
and other standard libraries (numpy, matplotlib, scipy, tqdm, etc.)
To install NEST version 3.7 you can follow instructions here. In Unix you can use conda and e.g., simply run
conda create --name nest_3_7_0 -c conda-forge nest-simulator=3.7.0=*
to create a new environment called nest_3_7_0 with installed the correct version of NEST.
We tested the network on a virtual machine with 16 CPUs and 16 GB of RAM. It takes
import nest
-- N E S T --
Copyright (C) 2004 The NEST Initiative
Version: 3.7.0
Built: May 24 2024 10:11:53
This program is provided AS IS and comes with
NO WARRANTY. See the file LICENSE for details.
Problems or suggestions?
Visit https://www.nest-simulator.org
Type 'nest.help()' to find out more about NEST.
import numpy as np
import math
import matplotlib.pyplot as plt
import scipy.stats as stats
from matplotlib import rcParams
import os.path
import scipy as scp
from scipy.interpolate import interp1d
import time
import os
from tqdm import tqdmfolder_name = 'inh_model_results/' # saving results to a folder
save = 1 # flag for saving results: 0 = off, 1 = save in folder: folder_name
N_CPU = 16 # Set according to the number of CPUs available in your machine
if save:
os.mkdir(folder_name)Functions¶
create_spectro(tone)¶
creates an entire spectrogram of time_sim length and 3500 frequency channels (same number as human IHCs population) the implemented spectrogram will present only a pure tone sound, stationary for the entire time_sim if gauss_on = 1, 21 channels will be activated (by setting a non-zero amplitude value), centred on the channel corresponding to the pure tone (channel_x) if gauss_on = 0, only channel_x will be activated with amplitude = 1
def create_spectro(tone):
channel_x = np.where(freq>=tone)[0][0]
spectro = np.zeros((3500,time_sim))
amplitudes = np.round(stats.norm.pdf(np.linspace(-1, 1, 21) , 0, 1.0/(math.sqrt(2*math.pi)*1)),2) #gaussian profile of amplitudes, with peak_amplitude = 1 for channel_x
if(gauss_on):
if(channel_x<10): #truncation of the gaussian profile of amplitudes
spectro[channel_x:channel_x+10+1,:] = amplitudes[10:].reshape(11,1)*np.ones((11, time_sim))
spectro[0:channel_x+1, :] = amplitudes[10-channel_x:11].reshape(channel_x+1,1)*np.ones((channel_x+1, time_sim))
else:
if(channel_x>3489): #truncation of the gaussian profile of amplitudes
spectro[channel_x-10:channel_x+1] = amplitudes[:11].reshape(11,1)*np.ones((11, time_sim))
spectro[channel_x:] = amplitudes[10:10+3500-channel_x].reshape(3500-channel_x,1)*np.ones((3500-channel_x, time_sim))
else:
spectro[channel_x - 10 : channel_x + 10 + 1, :] = amplitudes.reshape(21,1)*np.ones((21, time_sim))
else:
spectro[channel_x, :] = np.ones(time_sim)
return spectrocompute_ild_functions_ppg()¶
computes the functions that return an ILD value according to an azimuth angle, by interpolating “ild_values”, if PPG device are chosen
def compute_ild_functions_ppg():
x_values = np.array([-90,0,90])
if(ild_on):
y_values = ild_values
else:
y_values = np.repeat(ild_values[1],3)
r_function = interp1d(x_values, y_values, kind='linear')
l_function = interp1d(x_values[::-1], y_values, kind='linear')
"""fig, ax = plt.subplots(1)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.set_xlim(-100,100)
ax.set_xlabel("Angles [°]")
ax.set_ylabel("Spikes per Period")
ax.set_xticks(np.linspace(-90,90,7))
x = np.linspace(-90,90,181)
ax.plot(x, r_function(x), color = 'darkgreen', label = "Right Amplitude")
ax.plot(x, l_function(x), color = 'darkmagenta', label = "Left Amplitude")
ax.axhline(y = ild_values[1], xmin = 0.05, xmax = 0.95, color = 'r', label = "Mean Rate")
ax.legend()"""
return(r_function,l_function)compute_ild_functions_spg()¶
computes the functions that return an ILD value according to an azimuth angle, by interpolating “ild_rates”, if SPG device are chosen
def compute_ild_functions_spg():
x_values = np.array([-90,0,90])
if(ild_on):
y_values = ild_rates
else:
y_values = np.repeat(ild_rates[1],3)
def expfunc(x, a, b, c):
return a + (b * np.exp(c * x))
r_params, p_cov = scp.optimize.curve_fit(expfunc, x_values, ild_rates, bounds = ([-np.inf, 0, 0], [np.inf, np.inf, np.inf]))
l_params, p_cov = scp.optimize.curve_fit(expfunc, x_values, ild_rates[::-1], bounds = ([-np.inf, -np.inf, -np.inf], [np.inf, np.inf, np.inf]))
"""fig, ax = plt.subplots(1)
ax.set_xlim(-100,100)
ax.set_xlabel("Angles [°]")
ax.set_ylabel("Firing Rate [Hz]")
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.set_xticks(np.linspace(-90,90,7))
x = np.linspace(-90,90,181)
ax.plot(x, expfunc(x, *r_params), color = 'darkgreen', label = "Right Amplitude")
ax.plot(x,expfunc(x, *l_params), color = 'darkmagenta', label = "Left Amplitude")
ax.axhline(y = ild_rates[1], xmin = 0.05, xmax = 0.95, color = 'r', label = "Mean Rate")
ax.legend()"""
return r_params,l_paramscompute_itd(angle)¶
computes the value of ITD in milliseconds given the azimuth angle
def compute_itd(angle):
delta_x = (w_head*np.sin(np.deg2rad(angle)))
itd = 1000*delta_x/v_sound #ms
itd = np.round(itd,2) #only for spike_generators
return itdppg_set_up(spectro, ms)¶
sets up the Pulse Packet Generators (PPGs) according to the spectrogram, the ITD and the ILD functions
def ppg_set_up(spectro, ms):
for r in range(0, len(spectro)-1):
if spectro[r][ms] > 0:
r_ANFs_amp[10*r:10*(r+1)].set(pulse_times = np.around(np.arange(1, time_sim+1, 1000/freq[r]),2))
l_ANFs_amp[10*r:10*(r+1)].set(pulse_times = np.around(np.arange(1+itd, time_sim+itd+1, 1000/freq[r]),2))
"""print(freq[r], "Hz")
print(ms, "ms")
print(np.around(np.arange(1, time_sim+1, 1000/freq[r]),2))
print(np.around(np.arange(1+itd, time_sim+itd+1, 1000/freq[r]),2))"""
if ms in np.around(np.arange(0, time_sim, 1000/freq[r]), 0):
r_ANFs_amp[10*r:10*(r+1)].set(activity = int(spectro[r][ms]*r_function(angle)))
l_ANFs_amp[10*r:10*(r+1)].set(activity = int(spectro[r][ms]*l_function(angle)))
""""print("------------------------------")
print(int(spectro[r][ms]*r_num_spikes(angle)), "right activity")
print(int(spectro[r][ms]*l_num_spikes(angle)), "left activity")
print("------------------------------")"""
#ANF_noise to parrots
nest.Connect(ANFs_noise, r_ANFs[10*r:10*(r+1)], 'all_to_all')
nest.Connect(ANFs_noise, l_ANFs[10*r:10*(r+1)], 'all_to_all')spg_set_up(spectro, ms)¶
sets up the Sinusoidal Poisson Generators (SPGs) according to the spectrogram, the ITD and the ILD functions
def spg_set_up(spectro, ms):
def expfunc(x, a, b, c):
return a + (b * np.exp(c * x))
for r in range(0, len(spectro)-1):
if spectro[r][ms] > 0:
r_ANFs_amp[10*r:10*(r+1)].set(rate = ild_rates[1])
l_ANFs_amp[10*r:10*(r+1)].set(rate = ild_rates[1])
r_ANFs_amp[10*r:10*(r+1)].set(amplitude = spectro[r][ms]*expfunc(angle, *r_params))
l_ANFs_amp[10*r:10*(r+1)].set(amplitude = spectro[r][ms]*expfunc(angle, *l_params))
nest.Connect(ANFs_noise, r_ANFs[10*r:10*(r+1)], 'all_to_all')
nest.Connect(ANFs_noise, l_ANFs[10*r:10*(r+1)], 'all_to_all')Network Variables¶
General Variables¶
tones = [100] # [Hz], sound frequency of the pure tones tested
angles = np.arange(-90,100,15) #[°], range of azimuth angles
time_sim = 1000 #[ms]
w_head = 22 #[cm]
v_sound = 33000 #[cm/s]
ANF_device = 'PPG' #PulsePacket Generator vs Sinsoidal Poisson Generator 'SPG'
ild_on = 1 #[flag] for simulation with ILDs or only with ITDs in input
gauss_on = 1 #[flag] for pure tones stimulation, inclusion of lateral freqeuncy channels (i.e, for 100 Hz pure tones, we activate 21 channels with characteristic frequency between 99 and 101 Hz, centred in 100 Hz)
ild_values = [10,50,100] #[nr. of spikes], num of spikes for each pulse packet (PPG parameter)
sdev = 0.1 #[ms] Standard Deviation in PPG spikes for each pulse-packet (PPG parameter)
ild_rates = [100,125,300] #[Hz], rates for SPG
noise_rate = 0 # [Hz], valid for both typesNeuronal Populations Variables¶
#Single cells
V_m = -70 #mV
V_reset = -70 #mV
c_scb = 1
c_gcb = 1
c_mso = 1
c_lso = 1 #pF
#MSO cells
delays_mso = [1,1.3,1,0.45,0.44] #ms
mso_neurons = [1,2,3,4]
taus_mat = [[0.2, 0.1, 0.2, 0.01],
[0.2, 0.1, 0.5, 0.1],
[0.2, 0.1, 0.75, 0.14],
[0.2, 0.1, 1.2, 0.18]]
#taus90 [0.2, 0.1, 1.2, 0.18] # #taus50: [0.2, 0.1, 0.75, 0.14] #taus30: [0.2, 0.1, 0.5, 0.1] , #taus15: [0.2, 0.1, 0.2, 0.01]
#Synaptic Weights
ANFs2SBCs_weight = 16
ANFs2GBCs_weight = 8
GBCs2MNTBCs_weight = 16.0
MNTBCs2LSO_weight = -2.0
SBCs2LSO_weight = 8
SBCs2MSO_weight = 1
SBCs2MSO_inh_weight = [0,-30]
MNTBCs2MSO_weights = [0,-30]
n_battery = len(MNTBCs2MSO_weights) #implementation of 2 MSOs for each hemisphere, one with inhibitory inputs blocked
#Numerosity
n_IHCs = 3500
freq = np.round(np.logspace(np.log(20),np.log(20000), num = n_IHCs, base = np.exp(1)),2) #cochlea array of frequencies
n_ANFs = int(n_IHCs*10)
ANFs2SBCs = 4
ANFs2GBCs = 20
SBCs2MSOs = int(ANFs2GBCs/ANFs2SBCs)
SBCs2LSOs = int(ANFs2GBCs/ANFs2SBCs)
n_SBCs = int(n_ANFs/ANFs2SBCs)
n_GBCs = int(n_ANFs/ANFs2GBCs) #same of MNTB PCs and LSO PCs
n_MSOs = n_GBCs*n_battery #implementation of 2 MSOs for each hemisphere, one with inhibitory inputs blockedNetwork Simulation¶
start_time = time.time()
for tone in tones:
spectro = create_spectro(tone) #single tone
if(ANF_device == 'PPG'): #ILD functions depend on the ANF's model device chosen, they allow to compute the levels of ANFs firing rate used afterwards
r_function, l_function = compute_ild_functions_ppg()
else:
r_params, l_params = compute_ild_functions_spg()
results_r_MSO = np.zeros((n_battery, len(angles)))
results_l_MSO = np.zeros((n_battery, len(angles)))
results_r_LSO = np.zeros(len(angles))
results_l_LSO = np.zeros(len(angles))
for t in tqdm(range(len(taus_mat))):
taus = taus_mat[t]
mso_neuron_id = mso_neurons[t]
for angle in tqdm(angles):
nest.ResetKernel()
nest.set_verbosity('M_ERROR') # Suppress output for each simulation
nest.local_num_threads = N_CPU
nest.resolution = 0.01 # 10 us = minimum audible angle (MAA) increment of 1.25°
itd = compute_itd(angle)
if(ANF_device == 'PPG'):
r_ANFs_amp = nest.Create("pulsepacket_generator", n_ANFs,
params={"stop": time_sim, 'sdev': sdev})
l_ANFs_amp = nest.Create("pulsepacket_generator", n_ANFs,
params={"stop": time_sim, 'sdev': sdev})
if(ANF_device == 'SPG'):
ipds = 2*np.pi*itd*freq/1000
r_ANFs_amp = nest.Create('sinusoidal_poisson_generator',n_ANFs,
params={'frequency': np.repeat(freq, 10),'phase': np.repeat(np.rad2deg(ipds),10)}) #ITDs
l_ANFs_amp = nest.Create('sinusoidal_poisson_generator',n_ANFs,
params={'frequency': np.repeat(freq, 10),'phase': 0})
ANFs_noise = nest.Create('poisson_generator',1,
params = {'rate':noise_rate})
r_ANFs = nest.Create('parrot_neuron', n_ANFs)
l_ANFs = nest.Create('parrot_neuron', n_ANFs)
r_SBCs = nest.Create('iaf_cond_alpha', n_SBCs,
params = {'C_m': c_scb, 'V_reset': V_reset})
l_SBCs = nest.Create('iaf_cond_alpha', n_SBCs,
params = {'C_m': c_scb, 'V_reset': V_reset})
r_GBCs = nest.Create('iaf_cond_alpha', n_GBCs,
params = {'C_m': c_gcb, 'V_reset': V_reset})
l_GBCs = nest.Create('iaf_cond_alpha', n_GBCs,
params = {'C_m': c_gcb, 'V_reset': V_reset})
r_MNTBCs = nest.Create('iaf_cond_alpha', n_GBCs,
params = {'C_m': c_gcb, 'V_reset': V_reset})
l_MNTBCs = nest.Create('iaf_cond_alpha', n_GBCs,
params = {'C_m': c_gcb, 'V_reset': V_reset})
r_MSO = nest.Create('iaf_cond_beta', n_MSOs,
params = {'C_m': c_mso, 'tau_rise_ex' : taus[0], 'tau_rise_in' : taus[1], 'tau_decay_ex' : taus[2], 'tau_decay_in' : taus[3], 'V_reset': V_reset})
l_MSO = nest.Create('iaf_cond_beta', n_MSOs,
params = {'C_m': c_mso, 'tau_rise_ex' : taus[0], 'tau_rise_in' : taus[1], 'tau_decay_ex' : taus[2], 'tau_decay_in' : taus[3], 'V_reset': V_reset})
r_LSO = nest.Create('iaf_cond_alpha', n_GBCs,
params = {'C_m': c_lso, 'V_reset': V_reset})
l_LSO = nest.Create('iaf_cond_alpha', n_GBCs,
params = {'C_m': c_lso, 'V_m': V_m, 'V_reset': V_reset})
s_rec_r = nest.Create('spike_recorder')
s_rec_l = nest.Create('spike_recorder')
#Devices Connections
nest.Connect(r_ANFs, s_rec_r, 'all_to_all')
nest.Connect(l_ANFs, s_rec_l, 'all_to_all')
nest.Connect(r_SBCs, s_rec_r, 'all_to_all')
nest.Connect(l_SBCs, s_rec_l, 'all_to_all')
nest.Connect(r_GBCs, s_rec_r, 'all_to_all')
nest.Connect(l_GBCs, s_rec_l, 'all_to_all')
nest.Connect(r_MNTBCs, s_rec_r, 'all_to_all')
nest.Connect(l_MNTBCs, s_rec_l, 'all_to_all')
nest.Connect(r_MSO, s_rec_r, 'all_to_all')
nest.Connect(l_MSO, s_rec_l, 'all_to_all')
nest.Connect(r_LSO, s_rec_r, 'all_to_all')
nest.Connect(l_LSO, s_rec_l, 'all_to_all')
#ANFs
nest.Connect(r_ANFs_amp, r_ANFs, 'one_to_one')
nest.Connect(l_ANFs_amp, l_ANFs, 'one_to_one')
#ANF_parrots to SBCs
for i in range(n_SBCs):
nest.Connect(r_ANFs[ANFs2SBCs*i:ANFs2SBCs*(i+1)], r_SBCs[i], 'all_to_all', syn_spec = {"weight":ANFs2SBCs_weight})
nest.Connect(l_ANFs[ANFs2SBCs*i:ANFs2SBCs*(i+1)], l_SBCs[i], 'all_to_all', syn_spec = {"weight":ANFs2SBCs_weight})
#ANF_parrots to GBCs
for i in range(n_GBCs):
nest.Connect(r_ANFs[ANFs2GBCs*i:ANFs2GBCs*(i+1)], r_GBCs[i], 'all_to_all', syn_spec = {"weight":ANFs2GBCs_weight})
nest.Connect(l_ANFs[ANFs2GBCs*i:ANFs2GBCs*(i+1)], l_GBCs[i], 'all_to_all', syn_spec = {"weight":ANFs2GBCs_weight})
#GBCs to MNTBCs
nest.Connect(r_GBCs, r_MNTBCs, 'one_to_one', syn_spec = {"weight":GBCs2MNTBCs_weight, "delay": delays_mso[3]})
nest.Connect(l_GBCs, l_MNTBCs, 'one_to_one', syn_spec = {"weight":GBCs2MNTBCs_weight, "delay": delays_mso[3]})
#MSO
for i in range(n_GBCs):
for j in range(n_battery):
#Right MSO
#From SBCs (excitation):
nest.Connect(r_SBCs[SBCs2MSOs*i:SBCs2MSOs*(i+1)], r_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":SBCs2MSO_weight, "delay": delays_mso[0]}) #ipsilateral
nest.Connect(l_SBCs[SBCs2MSOs*i:SBCs2MSOs*(i+1)], r_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":SBCs2MSO_weight, "delay": delays_mso[2]}) #contralateral
#From LNTBCs (inhibition)
nest.Connect(r_SBCs[SBCs2MSOs*i:SBCs2MSOs*(i+1)], r_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":SBCs2MSO_inh_weight[j], "delay": delays_mso[1]}) #ipsilateral
#From MNTBCs (inhibition)
nest.Connect(l_MNTBCs[i], r_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":MNTBCs2MSO_weights[j], "delay": delays_mso[4]}) #contralateral
#Left MSO
#From SBCs (excitation):
nest.Connect(l_SBCs[SBCs2MSOs*i:SBCs2MSOs*(i+1)], l_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":SBCs2MSO_weight, "delay": delays_mso[0]}) #ipsilateral
nest.Connect(r_SBCs[SBCs2MSOs*i:SBCs2MSOs*(i+1)], l_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":SBCs2MSO_weight, "delay": delays_mso[2]}) #contralateral
#From LNTBCs (inhibition)
nest.Connect(l_SBCs[SBCs2MSOs*i:SBCs2MSOs*(i+1)], l_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":SBCs2MSO_inh_weight[j], "delay": delays_mso[1]}) #ipsilateral
#From MNTBCs (inhibition)
nest.Connect(r_MNTBCs[i], l_MSO[i*n_battery+j], 'all_to_all', syn_spec = {"weight":MNTBCs2MSO_weights[j], "delay": delays_mso[4]}) #contralateral
#LSO
for i in range(0, n_GBCs):
nest.Connect(r_SBCs[SBCs2LSOs*i:SBCs2LSOs*(i+1)], r_LSO[i], 'all_to_all', syn_spec = {"weight":SBCs2LSO_weight})
nest.Connect(l_SBCs[SBCs2LSOs*i:SBCs2LSOs*(i+1)], l_LSO[i], 'all_to_all', syn_spec = {"weight":SBCs2LSO_weight})
nest.Connect(r_MNTBCs, l_LSO, 'one_to_one', syn_spec = {"weight":MNTBCs2LSO_weight})
nest.Connect(l_MNTBCs, r_LSO, 'one_to_one', syn_spec = {"weight":MNTBCs2LSO_weight})
#Actual Simulation
for i in range(time_sim):
if(ANF_device == 'PPG'):
ppg_set_up(spectro,i)
nest.Simulate(1)
if(ANF_device == 'SPG'):
spg_set_up(spectro,i)
nest.Simulate(1)
#Data Collection
data_r = s_rec_r.get('events')
data_l = s_rec_l.get('events')
id_r_ANF1 = r_ANFs[0].get('global_id')
id_r_SBC1 = r_SBCs[0].get('global_id')
id_r_GBC1 = r_GBCs[0].get('global_id')
id_r_MNTBC1 = r_MNTBCs[0].get('global_id')
id_r_MSO1 = r_MSO[0].get('global_id')
id_r_LSO1 = r_LSO[0].get('global_id')
id_l_ANF1 = l_ANFs[0].get('global_id')
id_l_SBC1 = l_SBCs[0].get('global_id')
id_l_GBC1 = l_GBCs[0].get('global_id')
id_l_MNTBC1 = l_MNTBCs[0].get('global_id')
id_l_MSO1 = l_MSO[0].get('global_id')
id_l_LSO1 = l_LSO[0].get('global_id')
#LSO
rate_r_lso = len(data_r['times'][np.where(data_r['senders']>=id_r_LSO1)])
rate_l_lso = len(data_l['times'][np.where(data_l['senders']>=id_l_LSO1)])
ac_r_lso = np.unique(data_r['senders'][np.where((data_r['senders']>=id_r_LSO1))]) #active cells
ac_l_lso = np.unique(data_l['senders'][np.where((data_l['senders']>=id_l_LSO1))]) #active cells
n_ac_r_lso = len(ac_r_lso) #number of active cells
n_ac_l_lso = len(ac_l_lso) #number of active cells
#MSO
rate_r_mso = np.zeros(n_battery)
rate_l_mso = np.zeros(n_battery)
ac_r_mso = np.zeros((int(n_MSOs/n_battery),n_battery))
ac_l_mso = np.zeros((int(n_MSOs/n_battery),n_battery))
n_ac_r_mso = np.zeros(n_battery)
n_ac_l_mso = np.zeros(n_battery)
for i in range(int(n_MSOs/n_battery)): #n of batteries
for j in range(n_battery): # neurons for battery
if(id_r_MSO1+n_battery*i+j in data_r['senders']):
rate_r_mso[j] += (np.unique(data_r['senders'][np.where(data_r['senders'] == id_r_MSO1+n_battery*i+j)], return_counts= True)[1][0])
ac_r_mso[i,j] = id_r_MSO1 + i*n_battery + j #active cells
n_ac_r_mso[j] += 1 #number of active cells
else:
rate_r_mso[j] += 0
if(id_l_MSO1+n_battery*i+j in data_l['senders']):
rate_l_mso[j] += (np.unique(data_l['senders'][np.where(data_l['senders'] == id_l_MSO1+n_battery*i+j)], return_counts= True)[1][0])
ac_l_mso[i,j] = id_l_MSO1 + i*n_battery + j #active cells
n_ac_l_mso[j] += 1 #number of active cells
else:
rate_l_mso[j] += 0
#averaging on total number of active cells --> result: averege rate of the population
if((n_ac_r_mso[0] != 0) & (n_ac_l_mso[0] != 0) & (n_ac_r_mso[1] != 0) & (n_ac_l_mso[1] != 0) & (n_ac_r_lso != 0) & (n_ac_l_lso != 0)):
results_r_MSO[0, np.where(np.asarray(angles) == angle)[0][0]] = rate_r_mso[0]/n_ac_r_mso[0]
results_l_MSO[0, np.where(np.asarray(angles) == angle)[0][0]] = rate_l_mso[0]/n_ac_l_mso[0]
results_r_MSO[1, np.where(np.asarray(angles) == angle)[0][0]] = rate_r_mso[1]/n_ac_r_mso[1]
results_l_MSO[1, np.where(np.asarray(angles) == angle)[0][0]] = rate_l_mso[1]/n_ac_l_mso[1]
results_r_LSO[np.where(np.asarray(angles) == angle)[0][0]] = rate_r_lso/n_ac_r_lso
results_l_LSO[np.where(np.asarray(angles) == angle)[0][0]] = rate_l_lso/n_ac_l_lso
else:
results_r_MSO[0, np.where(np.asarray(angles) == angle)[0][0]] = rate_r_mso[0]
results_l_MSO[0, np.where(np.asarray(angles) == angle)[0][0]] = rate_l_mso[0]
results_r_MSO[1, np.where(np.asarray(angles) == angle)[0][0]] = rate_r_mso[1]
results_l_MSO[1, np.where(np.asarray(angles) == angle)[0][0]] = rate_l_mso[1]
results_r_LSO[np.where(np.asarray(angles) == angle)[0][0]] = rate_r_lso
results_l_LSO[np.where(np.asarray(angles) == angle)[0][0]] = rate_l_lso
if(save):
np.savetxt(folder_name + 'r_mso_results_tone_{}'.format(tone) + '_neuron_{}'.format(mso_neuron_id), results_r_MSO)
np.savetxt(folder_name + 'l_mso_results_tone_{}'.format(tone) + '_neuron_{}'.format(mso_neuron_id), results_l_MSO)
if(save):
np.savetxt(folder_name + 'r_lso_results_tone_{}'.format(tone), results_r_LSO)
np.savetxt(folder_name + 'l_lso_results_tone_{}'.format(tone), results_l_LSO)
end_time = time.time()
simulation_time = end_time - start_time
if(save):
np.savetxt(folder_name + 'simulation_time', [simulation_time]) #seconds 0%| | 0/4 [00:00<?, ?it/s]
0%| | 0/13 [00:00<?, ?it/s]
8%|▊ | 1/13 [04:31<54:19, 271.66s/it]
15%|█▌ | 2/13 [09:00<49:30, 270.01s/it]
23%|██▎ | 3/13 [13:29<44:53, 269.38s/it]
31%|███ | 4/13 [17:57<40:21, 269.05s/it]
38%|███▊ | 5/13 [22:25<35:50, 268.75s/it]
46%|████▌ | 6/13 [26:54<31:20, 268.64s/it]
54%|█████▍ | 7/13 [31:22<26:50, 268.41s/it]
62%|██████▏ | 8/13 [35:50<22:21, 268.23s/it]
69%|██████▉ | 9/13 [40:18<17:52, 268.13s/it]
77%|███████▋ | 10/13 [44:45<13:23, 267.87s/it]
85%|████████▍ | 11/13 [49:12<08:55, 267.68s/it]
92%|█████████▏| 12/13 [53:39<04:27, 267.47s/it]
100%|██████████| 13/13 [58:07<00:00, 268.24s/it]
25%|██▌ | 1/4 [58:07<2:54:21, 3487.11s/it]
0%| | 0/13 [00:00<?, ?it/s]
8%|▊ | 1/13 [04:27<53:28, 267.40s/it]
15%|█▌ | 2/13 [08:55<49:05, 267.76s/it]
23%|██▎ | 3/13 [13:22<44:32, 267.26s/it]
31%|███ | 4/13 [17:48<40:01, 266.81s/it]
38%|███▊ | 5/13 [22:14<35:31, 266.45s/it]
46%|████▌ | 6/13 [26:39<31:03, 266.24s/it]
54%|█████▍ | 7/13 [31:05<26:35, 265.96s/it]
62%|██████▏ | 8/13 [35:30<22:08, 265.74s/it]
69%|██████▉ | 9/13 [39:54<17:40, 265.06s/it]
77%|███████▋ | 10/13 [44:17<13:13, 264.41s/it]
85%|████████▍ | 11/13 [48:39<08:47, 263.96s/it]
92%|█████████▏| 12/13 [53:02<04:23, 263.54s/it]
100%|██████████| 13/13 [57:24<00:00, 265.00s/it]
50%|█████ | 2/4 [1:55:32<1:55:24, 3462.30s/it]
0%| | 0/13 [00:00<?, ?it/s]
8%|▊ | 1/13 [04:22<52:30, 262.55s/it]
15%|█▌ | 2/13 [08:45<48:09, 262.69s/it]
23%|██▎ | 3/13 [13:07<43:46, 262.66s/it]
31%|███ | 4/13 [17:31<39:27, 263.08s/it]
38%|███▊ | 5/13 [21:54<35:04, 263.11s/it]
46%|████▌ | 6/13 [26:17<30:41, 263.07s/it]
54%|█████▍ | 7/13 [30:41<26:18, 263.10s/it]
62%|██████▏ | 8/13 [35:04<21:56, 263.21s/it]
69%|██████▉ | 9/13 [39:27<17:32, 263.17s/it]
77%|███████▋ | 10/13 [43:51<13:09, 263.26s/it]
85%|████████▍ | 11/13 [48:15<08:47, 263.57s/it]
92%|█████████▏| 12/13 [52:38<04:23, 263.53s/it]
100%|██████████| 13/13 [57:03<00:00, 263.33s/it]
75%|███████▌ | 3/4 [2:52:35<57:24, 3444.52s/it]
0%| | 0/13 [00:00<?, ?it/s]
8%|▊ | 1/13 [04:23<52:39, 263.29s/it]
15%|█▌ | 2/13 [08:48<48:25, 264.10s/it]
23%|██▎ | 3/13 [13:11<43:59, 263.95s/it]
31%|███ | 4/13 [17:35<39:34, 263.85s/it]
38%|███▊ | 5/13 [21:58<35:09, 263.75s/it]
46%|████▌ | 6/13 [26:22<30:45, 263.66s/it]
54%|█████▍ | 7/13 [30:45<26:21, 263.54s/it]
62%|██████▏ | 8/13 [35:09<21:57, 263.49s/it]
69%|██████▉ | 9/13 [39:32<17:34, 263.54s/it]
77%|███████▋ | 10/13 [43:56<13:10, 263.51s/it]
85%|████████▍ | 11/13 [48:20<08:47, 263.73s/it]
92%|█████████▏| 12/13 [52:44<04:23, 263.68s/it]
100%|██████████| 13/13 [57:08<00:00, 263.69s/it]
100%|██████████| 4/4 [3:49:43<00:00, 3445.90s/it]
Plots¶
text_color = 'black'
rcParams['text.color'] = text_color
rcParams['axes.labelcolor'] = text_color
rcParams['xtick.color'] = text_color
rcParams['ytick.color'] = text_color
plt.rc('font', size=12) # controls default text sizes
plt.rc('axes', titlesize=16) # fontsize of the axes title
plt.rc('axes', labelsize=12) # fontsize of the x and y labels
plt.rc('xtick', labelsize=12) # fontsize of the tick labels
plt.rc('ytick', labelsize=12) # fontsize of the tick labels
plt.rc('legend', fontsize=12) # legend fontsize
plt.rc('figure', titlesize=16) # fontsize of the figure title
colors_l = ["magenta", 'crimson', "darkviolet", 'purple']
colors_r = ["green", 'darkgreen', 'forestgreen', "limegreen"]for tone in tones:
fig, ax = plt.subplots(1, 2, figsize=(15,5))
path_r = folder_name + 'r_lso_results_tone_{}'.format(tone)
path_l = folder_name + 'l_lso_results_tone_{}'.format(tone)
ax[1].set_title("Right LSO")
ax[0].set_title("Left LSO")
ax[1].plot(angles,np.loadtxt(path_r), 'go-')
ax[0].plot(angles,np.loadtxt(path_l),'mo-')
for i in range(len(angles)):
ax[0].axvline(angles[i], linewidth = 0.2, color = 'grey')
ax[1].axvline(angles[i], linewidth = 0.2, color = 'grey')
ax[0].set_xlabel("Angles [°]")
ax[0].set_ylabel("Mean Firing Rate [Hz]")
ax[0].set_xticks(angles)
ax[0].spines['top'].set_visible(False)
ax[0].spines['right'].set_visible(False)
ax[1].set_xlabel("Angles [°]")
ax[1].set_xticks(angles)
ax[1].spines['top'].set_visible(False)
ax[1].spines['right'].set_visible(False)
fig.tight_layout()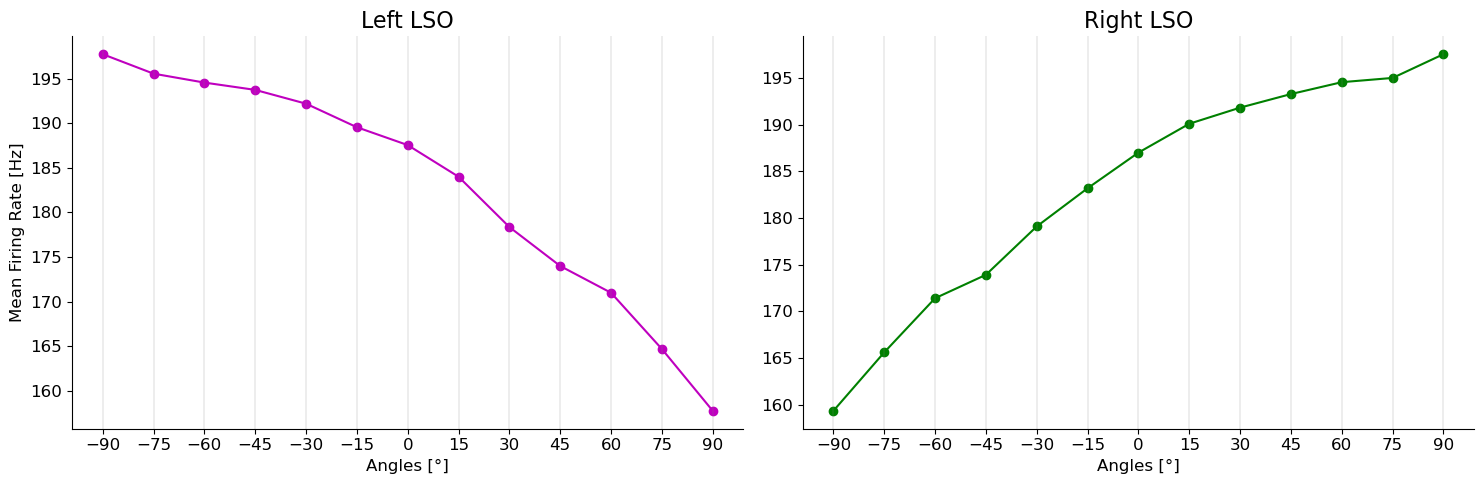
for tone in tones:
fig, ax = plt.subplots(1, 2, figsize=(15,5))
for mso_neuron_id in mso_neurons:
path_r = folder_name + 'r_mso_results_tone_{}'.format(tone) + '_neuron_{}'.format(mso_neuron_id)
path_l = folder_name + 'l_mso_results_tone_{}'.format(tone) + '_neuron_{}'.format(mso_neuron_id)
ax[1].set_title("Right MSO")
ax[0].set_title("Left MSO")
ax[1].plot(angles,np.loadtxt(path_r)[1], 'o-', color = colors_r[mso_neurons.index(mso_neuron_id)], label = "Neuron {}".format(mso_neuron_id))
ax[0].plot(angles,np.loadtxt(path_l)[1],'o-', color = colors_l[mso_neurons.index(mso_neuron_id)], label = "Neuron {}".format(mso_neuron_id))
for i in range(len(angles)):
ax[0].axvline(angles[i], linewidth = 0.2, color = 'grey')
ax[1].axvline(angles[i], linewidth = 0.2, color = 'grey')
ax[0].set_xlabel("Angles [°]")
ax[0].set_ylabel("Mean Firing Rate [Hz]")
ax[0].set_xticks(angles)
ax[0].legend(loc = 'lower left')
ax[0].spines['top'].set_visible(False)
ax[0].spines['right'].set_visible(False)
ax[1].set_xlabel("Angles [°]")
ax[1].set_xticks(angles)
ax[1].legend(loc = 'lower right')
ax[1].spines['top'].set_visible(False)
ax[1].spines['right'].set_visible(False)
fig.tight_layout()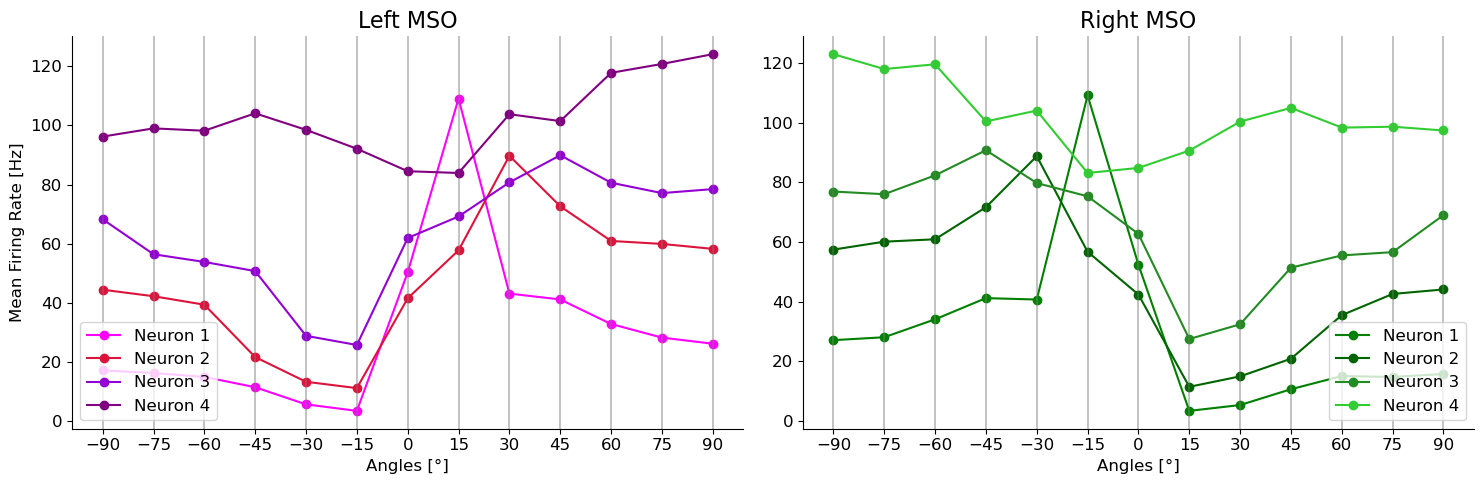
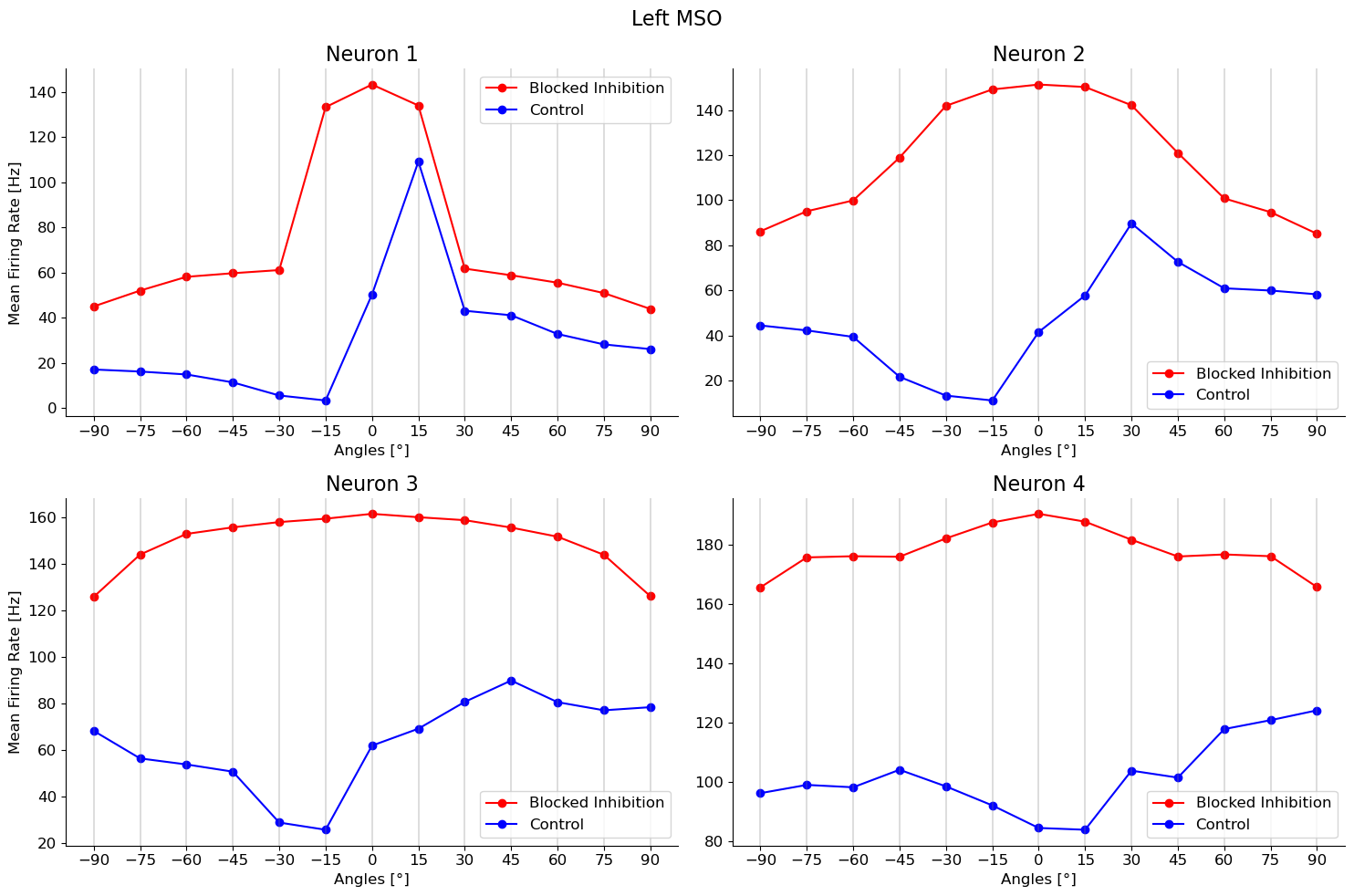
for tone in tones:
fig, ax = plt.subplots(2, 2, figsize=(15,10))
ax[0, 0].set_ylabel("Mean Firing Rate [Hz]")
ax[1, 0].set_ylabel("Mean Firing Rate [Hz]")
for mso_neuron_id in mso_neurons:
path_l = folder_name + 'l_mso_results_tone_{}'.format(tone) + '_neuron_{}'.format(mso_neuron_id)
fig.suptitle("Left MSO")
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].plot(angles,np.loadtxt(path_l)[0],"ro-", label = "Blocked Inhibition")
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].plot(angles,np.loadtxt(path_l)[1],"bo-", label = "Control")
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].set_title("Neuron {}".format(mso_neuron_id))
for i in range(len(angles)):
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].axvline(angles[i], linewidth = 0.2, color = 'grey')
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].axvline(angles[i], linewidth = 0.2, color = 'grey')
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].set_xlabel("Angles [°]")
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].set_xticks(angles)
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].legend()
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].spines['top'].set_visible(False)
ax[int(mso_neurons.index(mso_neuron_id)/2), mso_neurons.index(mso_neuron_id)%2].spines['right'].set_visible(False)
fig.tight_layout()